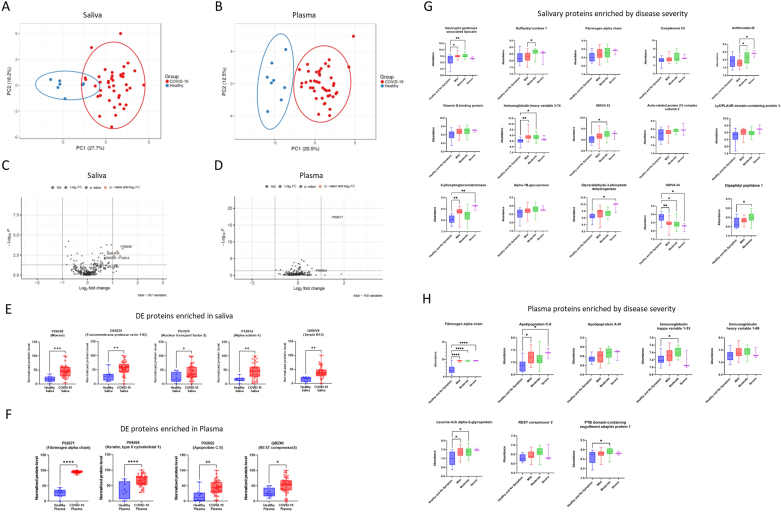
Fig. 3.
Comparative proteomic analyses revealed differentially expressed proteins (DEs) enriched in convalescent COVID-19 saliva and plasma. (A&B) Dimension reduction showed a separation of convalescent COVID-19 donors from healthy controls in saliva (A) and plasma (B). A nonparametric Wilcoxon rank sum test was applied to select subsets of proteins (p-value <0.01; 22 and 32 proteins were identified for saliva and plasma, respectively) before principal component analysis (PCA) was performed to visualize the samples. Circles indicate 95% confidence intervals of group memberships. Percentages along the axes indicate the degree of variance explained by that principal component. (C&D) Fold changes of protein expression of convalescent COVID-19 over healthy samples were plotted by negative log-transformed p-values in volcano plots. Dotted lines of the volcano plot represent thresholds for the fold changes (Log2 fold changes >1) and statistical significance (p < 0.05). (E&F) Differentially expressed proteins (Log2 fold changes>1, p < 0.05) in saliva and plasma, were depicted as in a relative abundance of five significantly up-regulated proteins. Individual dots represent individual values. The interquartile range, median, and min/max values were illustrated as box, middle line, and whiskers, respectively. Mixed-effect analysis with Tukey’s multiple comparisons test was used to measure statistical significance. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001. (G&H) Proteins differentially expressed (p < 0.05) by disease severity in saliva and plasma were depicted as in relative abundance. The interquartile range, median, and min/max values were illustrated as box, middle line, and whiskers, respectively. Mixed-effect analysis with Tukey’s multiple comparisons test was used to measure statistical significance. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.