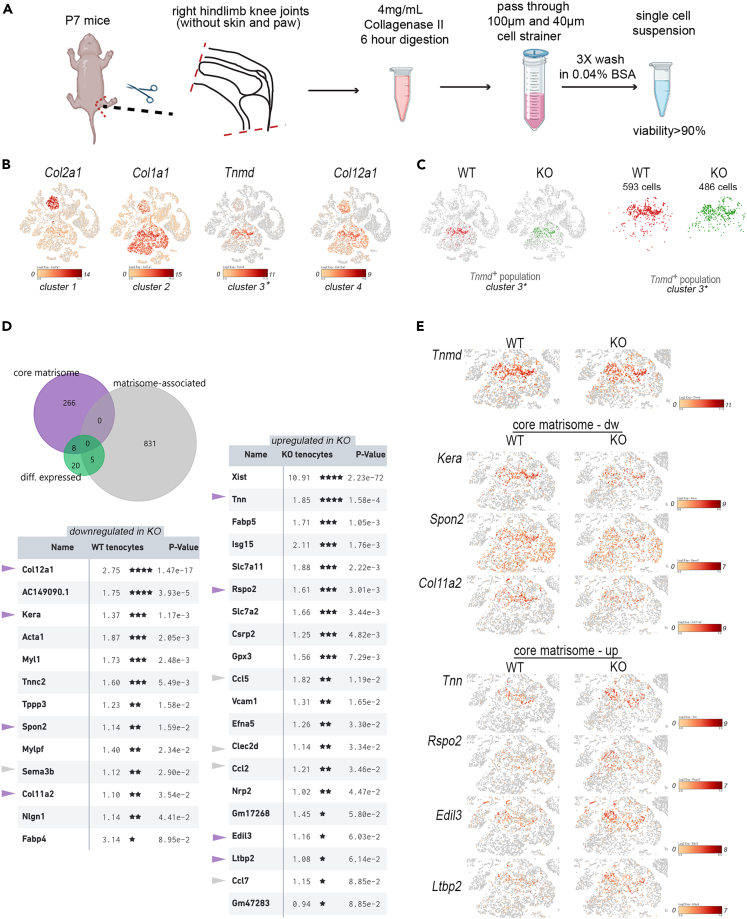
Figure 5.
Combined scRNA-seq and matrisome analysis of the Tnmd+ cell subpopulation
(A) Diagram of sample preparation for single cell sequencing. This figure is created with Biorender.com.
(B) Expression of cell type specific marker genes was used to annotate 4 different cell clusters. Log2 expression intensity values of individual genes are indicated in t-SNE plots (color scale).
(C) Tenocyte (Tnmd+) cell clusters (cluster 3) are shown in t-SNE plots and after gating.
(D) The proportions of differentially expressed entities within the core matrisome or matrisome associated cluster are shown in a Venn diagram. Genes that are down-or upregulated in tenocytes from Col12a1 KO are listed. Log2 fold changes between WT and Col12a1 KO mice and p values are shown. Entities of the core matrisome (purple arrowheads) or of the matrisome associated cluster (gray arrowheads) are highlighted. P- values were adjusted using the Benjamini-Hochberg correction for multiple tests ∗, p < 0.1; ∗∗, p < 0.05, ∗∗∗, p < 0.01; ∗∗∗∗, p < 0.001.
(E) Differentially expressed genes of the core matrisome within the Tnmd+ subpopulation are shown in t-SNE plots.