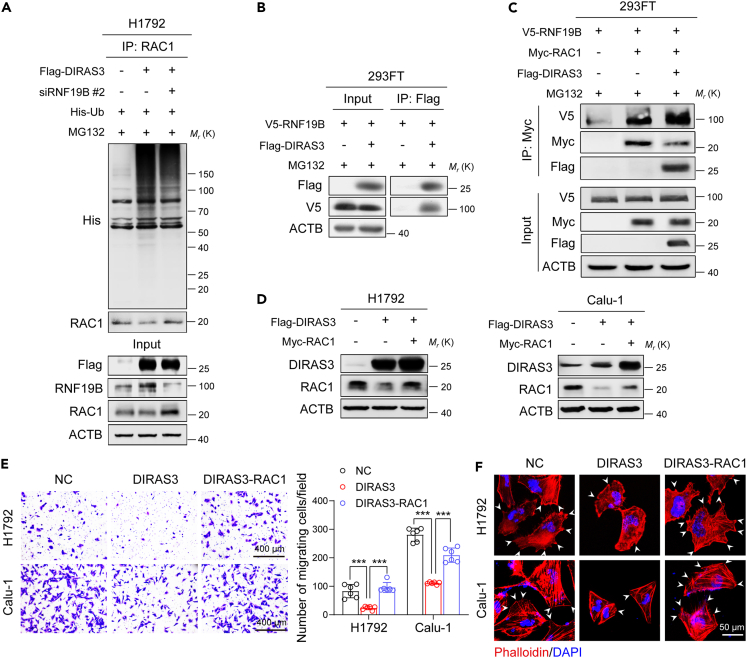
Figure 6.
The DIRAS3-RNF19B-RAC1 axis modulates migration of NSCLC cells
(A) Co-immunoprecipitation analysis of the ubiquitination of RAC1 in H1792 cells co-transfected with Flag-DIRAS3 plasmid, RNF19B-2 siRNA, and His-Ub plasmid.
(B) 293FT cells were transfected with V5-RNF19B and Flag-DIRAS3 plasmids and were cultured for 24 h before being further incubated with MG132 (20 μM) for 6 h. Then, the co-immunoprecipitation assays were carried out with Flag antibody and the co-eluted proteins were detected by western blot assays with Flag and V5 antibodies.
(C) 293FT cells were transfected with V5-RNF19B, Myc-RAC1, and Flag-DIRAS3 plasmids and were cultured for 24 h before being further incubated with MG132 (20 μM) for 6 h. Then, the co-immunoprecipitation assays were carried out with Myc antibody and the co-eluted proteins were detected by western blot assays with V5, Myc, and Flag antibodies.
(D) Overexpression of DIRAS3 or RAC1 in H1792 and Calu-1 cells. Cell lysates were analyzed by western blotting with antibodies against DIRAS3, RAC1, and ACTB.
(E) Transwell assays of H1792 and Calu-1 cells that overexpressed DIRAS3 and RAC1. The statistics were performed using two-tailed Student’s t-tests, ∗∗∗p < 0.001. (F) TRITC-phalloidin staining of H1792 and Calu-1 cells that overexpressed DIRAS3 and RAC1. The white arrows denote the typical protrusions.