Figure 4.
BiFCPL enables biotin labeling of proteins at mitochondria-LD contact sites
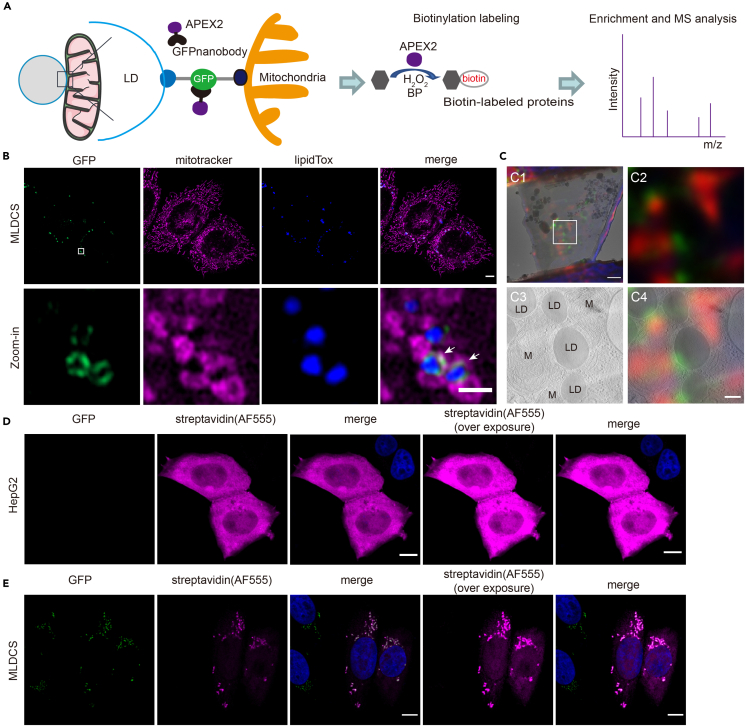
(A) Designing principle to characterize the spatial proteome at mitochondria-LD contacts.
(B) Representative single plane images of the reporter cell line (MLDCS) stably expressing mito-sfGFP11 and sfGFP1-10-Plin2. MitoTracker and LipidTox were used to label mitochondria and LDs, respectively. Scale bar, 5 μm. The lower images are a zoom-in version of the boxed region in the upper images. The white arrows mark contact sites between mitochondria and LDs. Scale bar, 1 μm. See also Figure S5. Contact, green; mitochondria, magenta; LDs, blue.
(C) Correlative light microscopy (LM) and electron microscopy (EM) analysis of the MLDCS cells. C1, superimposition of the LM image and EM image of the prepared lamella; C2-4, LM image (C2), EM image (C3), and correlation of LM and EM image (C4) of the boxed region in C1. Scale bar, 1 μm in C1 and 200 nm in C4. Contact, green; mitochondria, red.
(D and E) Representative fluorescence images of the wild-type HepG2 cells (D) or MLDCS cells. The cells were transiently transfected with GNAP2 and then subjected to fluorescence labeling after biotin labeling. Alexa 555-conjugated streptavidin was used to detect the biotin-labeled proteins. Scale bar, 5 μm. Contact, green; streptavidin, magenta; nucleus, blue.