Figure 1.
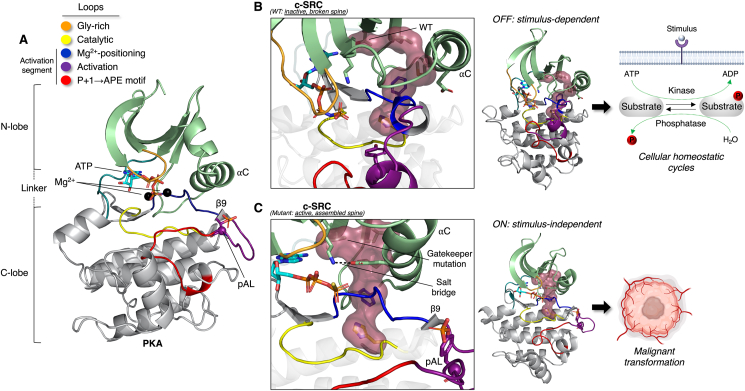
Typical architecture of a eukaryotic protein kinase (ePK) and structural considerations in cancer. A) PKA (PDB: 1ATP) is displayed as the prototypical model of an ePK. The catalytic core is defined by N- and C-terminal lobes separated by a linker that creates an active site cleft accommodating ATP and two divalent cations (Mg2+). The N-lobe consists of a five-stranded β-sheet and mobile αC helix (note, the preceding αB helix is not always present in ePKs), while the C-lobe is predominantly helical, typically α-helices D through to I. The major conserved loops and motifs, from N- to C-termini, are indicated in the colour-coded key and this colour scheme remains consistent throughout the manuscript. Briefly, the glycine (Gly)-rich loop connecting the first two β-strands of the N-lobe helps position ATP for catalysis. All subsequent regions of interest are located in the C-lobe. The catalytic loop contains a critical HRD (His-Arg-Asp) motif involved in phospho-transfer and stabilisation of the activation loop. The DFG motif is also referred to as the magnesium-positioning loop. A feature of an active kinase is also a small, β9-strand formed between the DFG motif and activation loop. As touched upon, the activation loop contains the important phosphorylation site (‘pAL’ in the figure) that is a signature of most active protein kinases (e.g., the basic HRD motif arginine interacts with the negatively charged phosphate group). Lastly, the P+1 loop and APE motif generally help position a substrate during the catalytic cycle. B) Several residues from these loops contribute to an internal regulatory ‘spine’ that is disrupted in the inactive kinase (presented as surface representation). In the case of the tyrosine kinase c-SRC (PDB: 2SRC), this is due to a threonine residue occupying the so-called gate-keeper position in the 5th β-stand of the N-lobe, ensuring fidelity of phospho-turnover cycles inherent to cellular homeostasis. C) Substitution of this residue to a bulkier hydrophobic amino acid (in this case isoleucine; PDB: 3DQW) results in constitutive spine formation and kinase activation, as evidenced by several other features including, but not limited to, pAL, formation of the β9 strand, and a salt bridge between an αC helix glutamate with a lysine in the β3-strand. Without the normal homeostatic restraints placed on activating an ePK, this single mutation is sufficient to trigger malignant transformation.