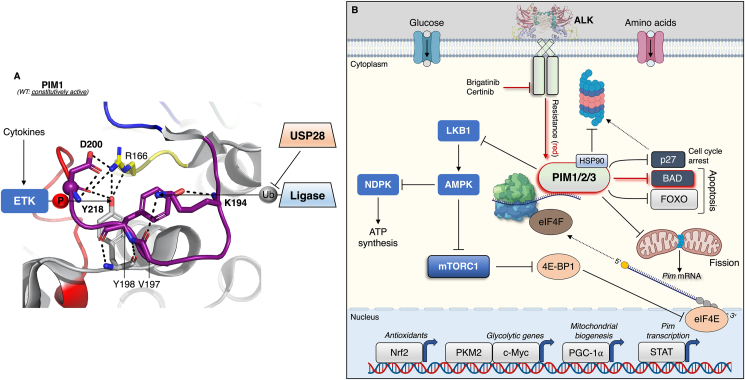
Figure 3.
Regulation of PIM kinase activity and considerations for its role in neuroblastoma. A) PIM is synthesized as a constitutively active enzyme devoid of activation loop phosphorylation attributable to a phospho-mimetic D200 (numbering based on PIM1; PDB: 1XR1) at the canonical phospho-acceptor position. While phosphorylation is not absolutely required for activity, Y218, a substrate of ETK, albeit in response to cytokine stimulation, participates in the extensive hydrogen bonding network associated with the active enzyme. K194, a putative ubiquitination substrate, is also implicated in this network, and may affect protein stability via ubiquitin turnover (e.g., USP28-mediated deubiquitination). B) The global effects of the PIM kinase signalling milieu are an upregulation of metabolic processes that maximise energy extraction (e.g., PGC-1α-induced mitochondrial biogenesis), protection of the cells against oxidative stress (e.g., Nrf2-induced antioxidant defence) and progression through the cell cycle (inhibition of apoptotic mediators and p27). PIM expression is sensitive to nutrient status, probably through activation of the master growth regulator mTORC1, and as such may promote ATP synthesis via inhibition of AMPK. Like what was presented for ALK in Figure 3C, drug resistance implicating activation of PIM and downstream signalling is indicated in red.