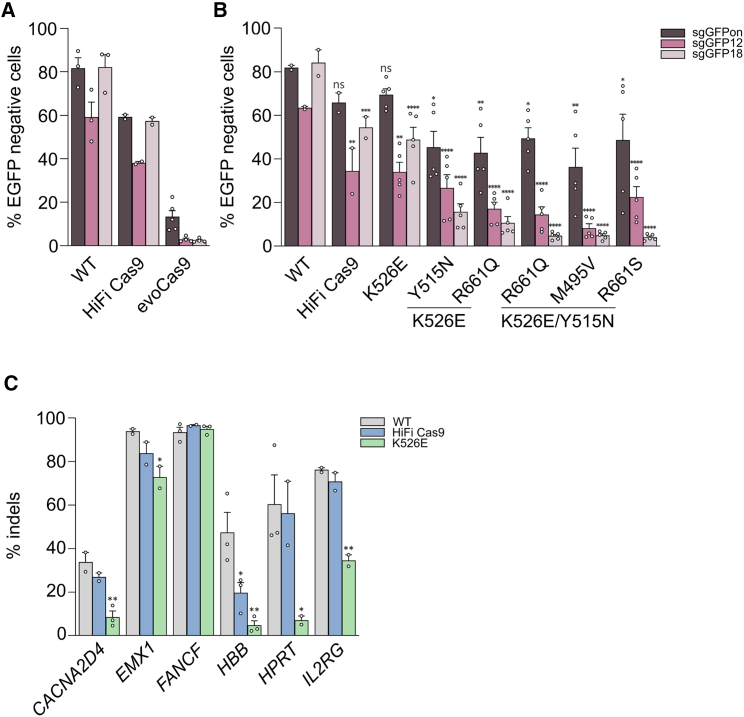
Figure 1.
Editing activity and precision of RNP from SpCas9 high-fidelity variants
(A and B) HEK293T cells stably expressing EGFP were lipofected with the indicated RNPs together with sgRNA perfectly matching the target (sgGFPon) or containing a single mismatch at different positions along the spacer as indicated (sgGFP12 and sgGFP18: mismatched nucleotides in positions 12 and 18 counting from the PAM, respectively). Loss of fluorescence was measured by cytofluorimetry 7 days post-lipofection. In (B), statistical significance was assessed using one-way ANOVA, comparing each mutant with WT SpCas9 separately for sgGFPon, sgGFP12, and sgGFP18. (C) Editing activity (percentage of indels) measured by sequencing (tracking indels by deconvolution [TIDE]) at the indicated endogenous genomic loci in U2OS cells 3 days after electroporation of WT SpCas9, HiFi Cas9, or K526E RNPs. Statistical significance was assessed using one-way ANOVA, comparing HiFi Cas9 and K526E with WT SpCas9. Data reported as mean ± SEM for n ≥ 2 biologically independent replicates.