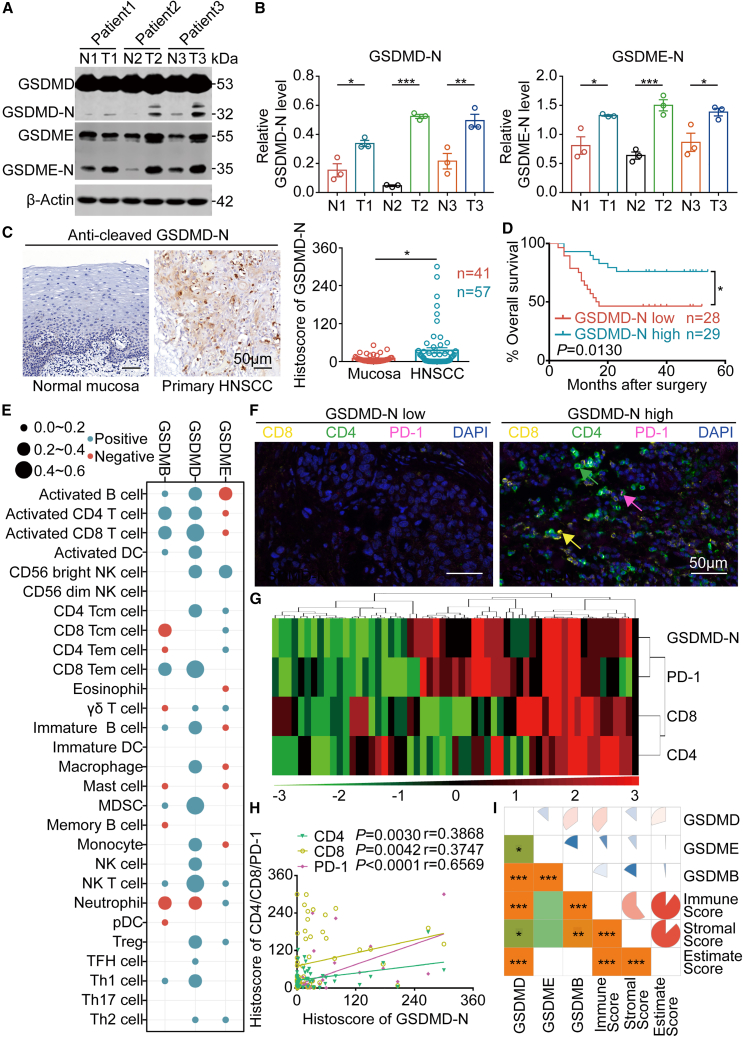
Figure 1.
Expression of GSDM family proteins in HNSCC
(A and B) The production of GSDMD-N and GSDME-N in paired normal oral mucosal tissue (N) and HNSCC (T) was measured using western blotting (left) and corresponding quantitative analysis (right). (C) Representative immunochemistry images of GSDMD-N production in primary HNSCC and normal mucosal tissues (left). Histoscores of GSDMD-N determined from GSDMD-N production levels in normal mucosal (n = 41) and primary HNSCC (n = 57) tissue samples (right). Scale bars, 50 μm. (D) Kaplan-Meier (K-M) survival analysis of the HNSCC patient cohort divided into high and low GSDMD-N production groups. (E) Bubble chart of the correlations between GSDM family proteins (GSDMB, GSDMD, and GSDME) and ssGSEA of 28 immune cell types in HNSCC samples from TCGA. (F) Representative graphs of multiplex immunohistochemistry (IHC; CD8, CD4, PD-1, and DAPI) in HNSCC patient cohort sections (high GSDMD-N production vs. low GSDMD-N production). CD8: yellow; CD4: green; PD-1: pink; DAPI: blue. Scale bars, 50 μm. (G) Positive correlations among CD8, CD4, PD-1, and GSDMD-N in the HNSCC microenvironment are displayed by hierarchical clustering. The color scale represents the histoscore. (H) The correlations between CD8, CD4, PD-1, and GSDMD-N in the HNSCC microenvironment. (I) Spearman’s correlation analysis between GSDM family proteins (GSDMB, GSDMD, and GSDME) and the immune score, stromal score, and ESTIMATE score. Data, mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.