Figure 4.
CRISPRa and CRISPRi pooled screens
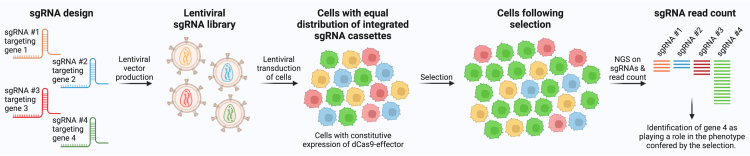
Pooled CRISPRa or CRISPRi screens begin with designing the sgRNA library, which can be genome-wide or target a subset of genes, for example transcription factors or a custom gene panel. This simple example contains four different sgRNAs each targeting a different gene. Lentiviral plasmids are cloned containing the sgRNA expression cassette and then lentiviral vectors are produced and used to transduce cells that usually already constitutively express the dCas9-effector that facilitates gene modulation. Transduction is performed at a low multiplicity of infection ensuring that only a single sgRNA expression cassette is integrated in each cell. The vector also contains a selection marker enabling depletion of untransduced cells. Throughout the process, it is important to confirm abundant and similar representation of sgRNAs. Next, a selection is applied to the cells either by means of increased or decreased survival/proliferation or FACS-based selection based on a specific phenotype or function. This usually follows a period of exposure to a stimulus such as drug treatment. Following selection, sgRNA cassettes are sequenced and quantified by NGS, and selection for or against a specific sgRNA signifies an involvement of the sgRNA’s target gene in the process that was investigated, for example drug resistance.