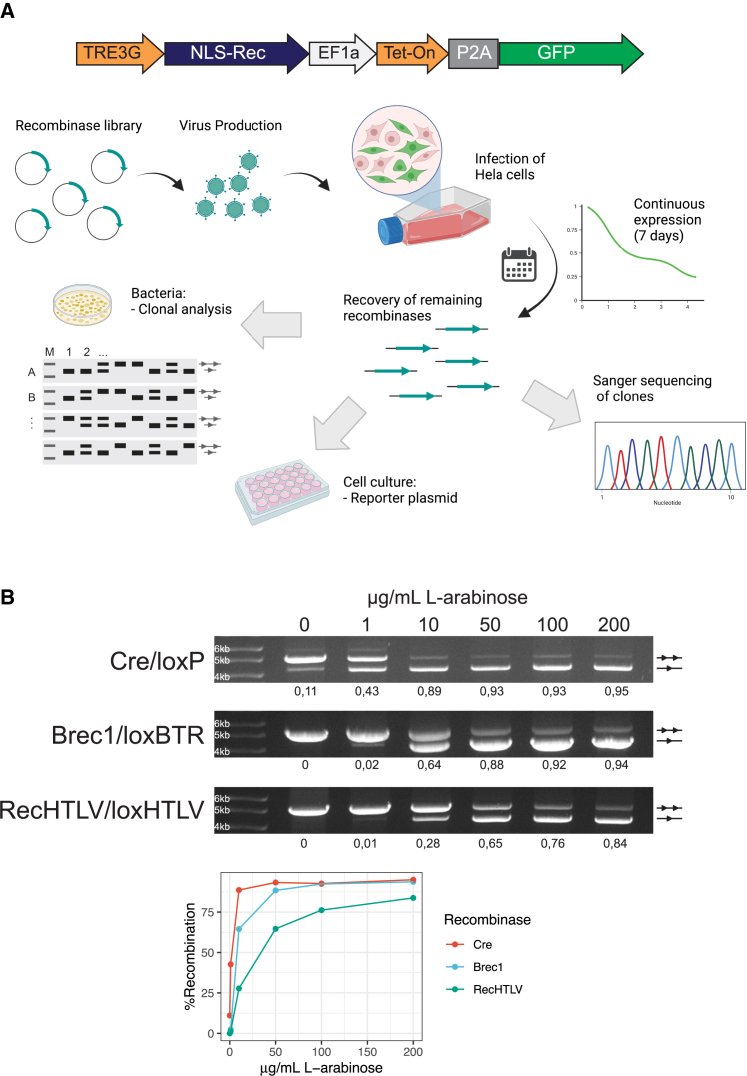
Figure 2.
Screening for an active and specific HTLV-1 recombinase
(A) Overview of the process for the selection of clones. In the upper part, a scheme of the construct used for expression of the library is shown: expression of the recombinase fused to a nuclear localization signal (NLS-Rec) is regulated by a tetracycline-inducible promoter (TRE3G), which is bound by Tet-On 3G (Tet-On advanced transactivator protein) upon doxycycline treatment; bicistronic expression of Tet-On 3G and a GFP cassette is driven by an elongation factor 1 alpha (EF1a) promoter and separated by a self-cleaving 2A peptide (P2A). The lower part shows the process followed for selection of candidate clones: the recombinase library was cloned into a lentiviral vector and, after lentivirus production, HeLa cells were infected with the recombinase-containing lentiviral particles. After 7 days of expression, the recombinases were extracted by PCR and cloned in the pEVO bacterial plasmid for sequencing and evaluation of activity on loxHTLV. Furthermore, the clones were tested in the cell culture for activity on the loxHTLV target site. (B) Top: agarose gels for the plasmid activity assay showing recombination efficiency of Cre, Brec1, and RecHTLV on their respective target sites in bacteria. The performed test digest results in a smaller fragment for recombined (one triangle) and a larger fragment for non-recombined plasmids (two triangles). The recombinases were tested at 0, 1, 10, 50, 100, and 200 μg/mL L-arabinose as shown above. Quantification of recombination is shown below the gels as a fraction of 1 (1 fully recombined, 0 not recombined). Bottom: line graph showing the quantification of recombination at the different levels of arabinose for Cre, Brec1, and RecHTLV. Parts of the figure were created with BioRender.com.