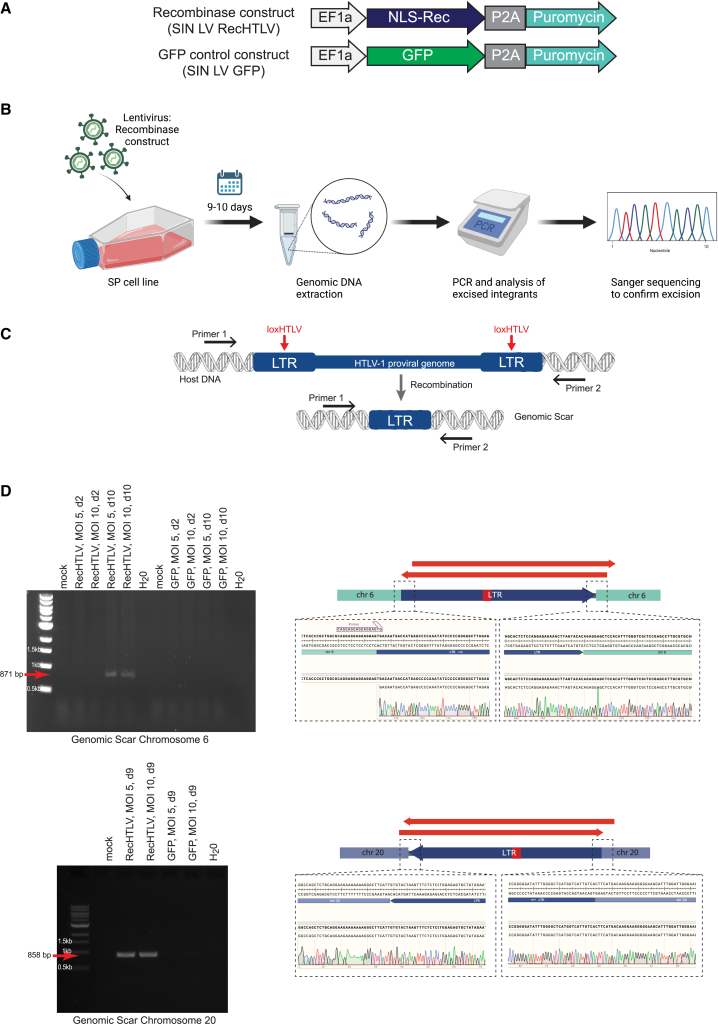
Figure 8.
RecHTLV expression excises the integrated HTLV-1 provirus from infected cells
(A) Schematic representation of the used lentiviral expression constructs SIN LV RecHTLV and SIN LV GFP. The EF1a promoter drives the expression of the RecHTLV, the HTLV-1 specific recombinase fused to nuclear localization signal (NLS-Rec) or GFP, followed by a P2A peptide, which allows the expression of a puromycin resistance gene. (B) Experimental workflow: the HTLV-1-infected SP cell line was transduced with lentiviral vectors expressing RecHTLV or GFP. After 9–10 days, genomic DNA was extracted and a PCR was performed followed by Sanger sequencing to confirm the correct excision of the HTLV-1 provirus. (C) Scheme of the HTLV-1 provirus integrated into the genome of the SP cells. The black arrows indicate the position of the primers used, which bind in the genomic DNA of the host cell. Upon recombination on the loxHTLV sites by RecHTLV, the proviral genome will be excised leaving a “genomic scar” consisting of one LTR sequence. (D) Left: PCR on genomic DNA of recombinase-treated SP cells with primers as indicated in (B) for the proviruses in chromosomes 6 and 20. Marked with red arrows are the PCR products, which indicate excision of proviral DNA. MOI indicates multiplicity of infection of the recombinase or GFP constructs and d indicates the days post transduction. Right: representation of the sequencing strategy and Sanger sequencing results from the genomic scar on chromosomes 6 and 20 following RecHTLV expression. The red arrows indicate the extent of the Sanger sequencing reads, which confirm the correct excision product. Parts of the figure were created with BioRender.com.