Figure 2.
Ex vivo detection and analysis of mouse implanted human neural progenitor cells in coronal brain slices
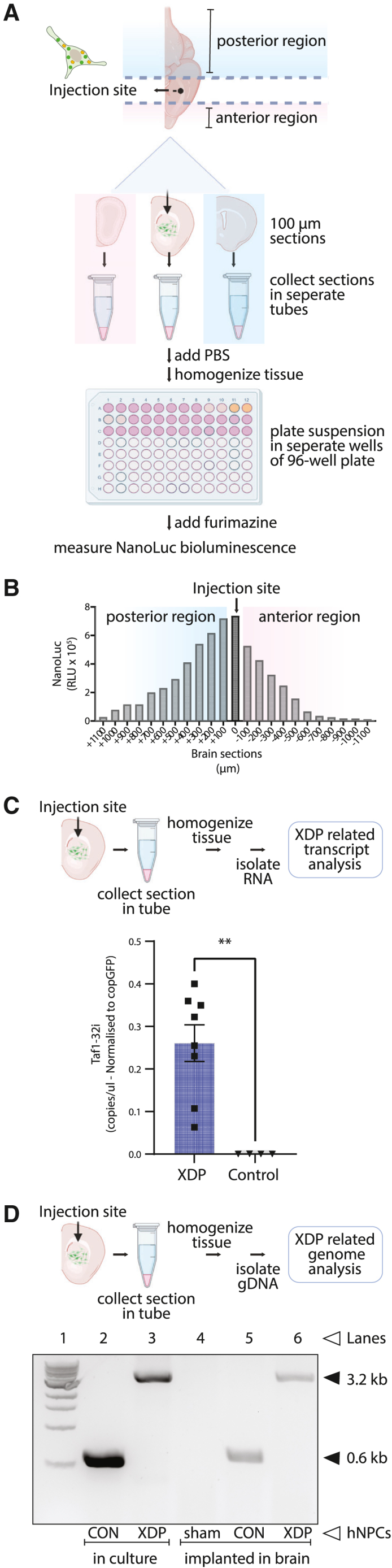
(A) Schematic illustrating the procedure for hNPC analysis post-implantation in mouse brains. Frozen brain slices (100-μm thickness) were collected in the coronal plane in the left hemisphere around the injection site and placed in microcentrifuge tubes. NanoLuc bioluminescence levels in each tube indicated the brain slices that contained the injected hNPCs. (B) Detection of hNPCs in serial brain slices. A peak of NanoLuc luminescence generated by ENoMi-expressing hNPCs corresponded to the region around the injection site. The injection site is annotated as 0 μm. (C) Schematic explaining how levels of TAF1-32i transcript in the implanted area were measured by ddPCR. Measurements were performed on RNA isolated from brain slices with NanoLuc bioluminescence derived from implanted ENoMi-expressing XDP hNPCs (n = 8) or control hNPCs (n = 4). Values and error bars reflect mean ± SEM. Data were analyzed with an unpaired t test. ∗∗p < 0.01. (D) Schematic demonstrating how to detect the SINE-VNTR-Alu (SVA) insertion in the TAF1 gene of implanted hNPCs derived from XDP patients. This procedure was performed hNPCs in culture and implanted in brain slices of mice. The genomic SVA-insertion region was PCR amplified with primers and resolved by gel electrophoresis to confirm the presence of the SVA in XDP hNPCs. First Lane: 1 kb DNA ladder. Second Lane: hNPCs from non-XDP hNPCs in culture. Third Lane: XDP hNPCs in culture. Fourth Lane: Collected brain slices from a sham (PBS)-injected mouse. Fifth Lane: Brain slice containing non-XDP hNPCs. Sixth Lane: Brain slice containing XDP hNPCs. The predicted 3,229-bp SVA product was present in all XDP samples (upper arrow), whereas hNPCs of healthy individuals (control) had a product of ∼599 bp (lower arrow), a difference consistent with the size of this SVA.
