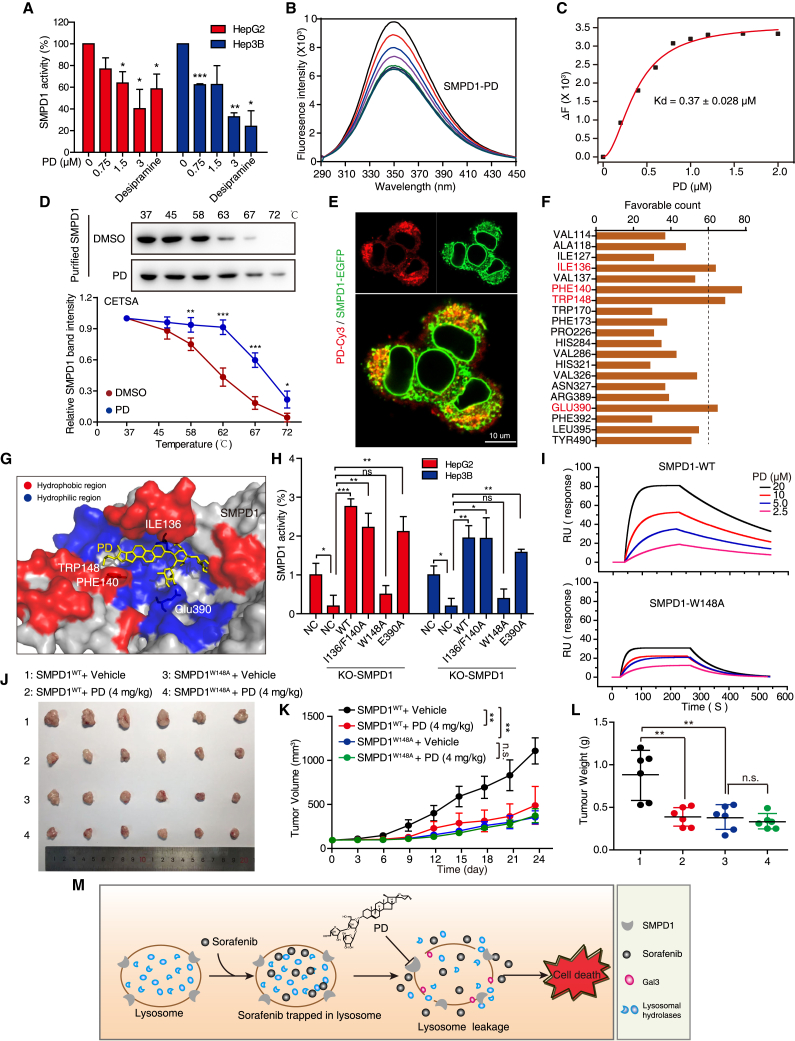
Figure 8.
PD inhibits HCC cell growth by targeting the surface groove of SMPD1 at W148
(A) Enzymatic activity of SMPD1 in HCC cells treated with increasing concentrations of PD (0–3 μM) or desipramine (10 μM, positive control) was determined by SMPD1 activity assay. (B) The interaction between PD and SMPD1 was determined by fluorescence titration experiments. (C) The fluorescence quenching of SMPD1 at 340 nm vs. the PD concentration was fitted with Hill plot. (D) CETSA curves comparing the change in the thermal stability of recombinant SMPD1 upon treatment with PD and DMSO. (E) Confocal microscope of cells expressing SMPD1-GFP after treatment with PD-Cy3 (1 μM), the colocalization of SMPD1 and PD-Cy3 was imaged. Scale bar, 10 μm. (F) The favorable counts for each amino acid of SMPD1 bound with PD. (G) Proposed binding pose depiction of the potential interaction mode between PD and SMPD1. Red, hydrophobic region; blue, hydrophilic region. (H) SMPD1WT, SMPD1I136/P140A, SMPD1W148A, or SMPD1E390A was introduced into SMPD1-KO cells, and the enzymatic activity of SMPD1 was detected by SMPD1 activity assay. (I) SPR sensorgram for the association of PD with immobilized SMPD1WT and SMPD1W148A. (J–L) Tumors formed by HepG2-SMPD1-KO cells expressing WT or W148 mutant SMPD1 proteins were treated with or without PD (i.g 4 mg/kg), then the tumor photos (J), tumor growth curves (K), and tumor weight (L) were compared (n = 6). All data are represented as the mean ± SD; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (M) Schematic diagram summarizing how PD overcomes sorafenib resistance in HCC.