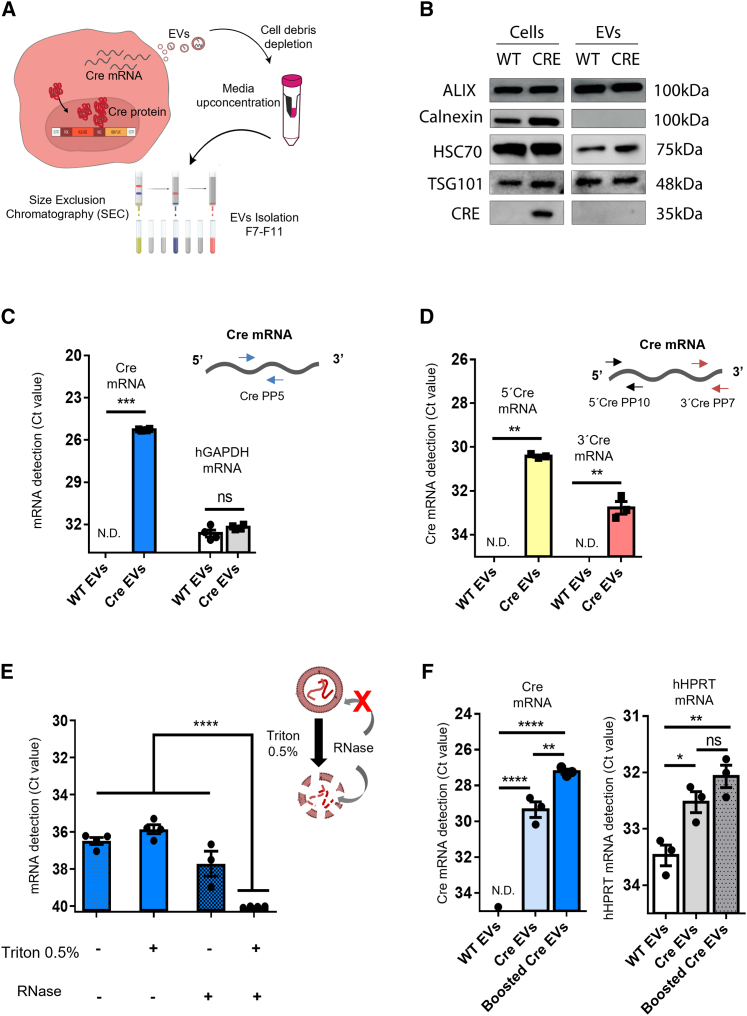
Figure 2.
Cre activity is mediated by transfer of Cre mRNA through EVs
(A) Schematic representation of EV isolation by size exclusion chromatography (SEC). In brief, EVs were isolated from the medium of HEK293T stably expressing Cre, cell debris was removed (300 × g × 10 min) and medium concentrated (100 kDa filter) to a final volume of 500 μL and then loaded onto a qEV Original SEC column. Five EV-enriched fractions of 500 μL were collected (fractions 7–11). (B) Western blotting of equimolar amounts of protein from cells and their derived EVs shows the positive markers Alix, HSC70, and TSG101 and undetectable levels of the ER marker calnexin. Cre protein is present in Cre donor cells but was not detectable in EVs from those cells. (C) Cre mRNA is detected in Cre EVs, but not WT EVs (n = 4). hGAPDH was detected in both conditions. Data are presented as Ct values, mean ± SEM and compared by unpaired t test with Welch’s correction. ∗∗∗p < 0.001; ns, not significant. (D) 5′ and-3′ regions of Cre exRNA are detected in Cre EVs, but not in WT EVs (n = 3). Data are presented as Ct values, mean ± SEM and compared by unpaired t test with Welch’s correction. ∗∗p < 0.01. (E) Cre EVs treated with RNase A in the presence or absence of 0.5% Triton X-100 showed that Cre-exRNA is predominantly protected inside EVs (n = 4). Data are presented as mean ± SEM and compared by ordinary one-way ANOVA followed by Dunnett’s multiple comparison test (F = 493.4). ∗∗∗∗p < 0.0001. (F) CMV-STEAP3-SDC4-NadB booster plasmid increases EV production and Cre exRNA detection. hHPRT was used as a housekeeping control. Data are presented as mean ± SEM and compared by ordinary one-way ANOVA followed by Sidak’s multiple comparisons test (F = 192.4). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, not significant.