Extended Data Fig. 6. AlphaFold2 analysis of TMEM164 active site cavity.
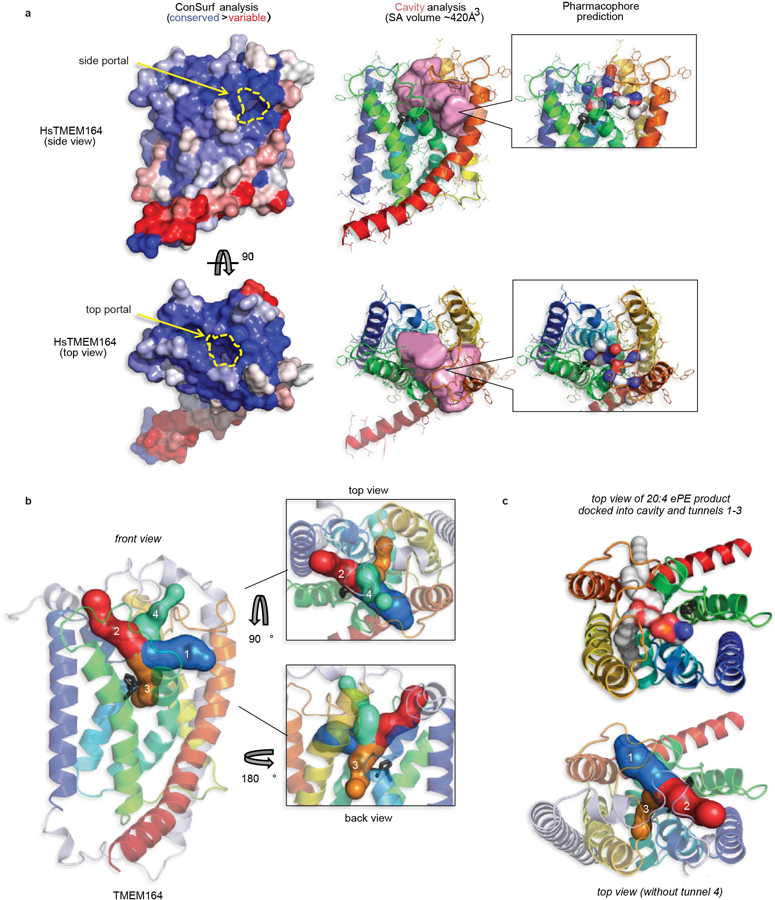
a, The human TMEM164 structural model was processed by the ConSurf server (http://consurf.tau.ac.il) to map the sequence conservation profile of its underlying family to the 3D fold surface color-ramped from blue (conserved) to red (variable) patches. For side and top views, the ConSurf surfaces are aligned to the ribbon model of TMEM164 embedded with the CavityPlus-calculated volume in pink (server at http://pkumdl.cn), showing the positions of predicted portals to the lipid bilayer. In addition, the CavPharmer tool at CavityPlus was used to predict a pharmacophore skeleton for a prospective ligand (which could represent the lipid substrates or product of the enzymatic reaction) for TMEM164, showing a shape and chemical nature in agreement with a predicted function as a C20:4-preferring lyso-ePL acyltransferase. b, The program CAVER (https://loschmidt.chemi.muni.cz/caverweb) was used to visualize the tunnels leading from the central TMEM164 cavity that abuts the Cys123/His181 predicted catalytic dyad (shown in black stick format), to the portals that open to the lipid bilayer (tunnels 1 and 3, respectively in blue and orange, with lengths 12.8 Å and 16.7 Å) and to the outer leaflet of the membrane (tunnels 2 and 4, respectively in red and green, both with lengths of 15.1 Å). c, A top view of the six-transmembrane core of TMEM164 with docked C20:4 ePE, with acyl chains occupying tunnels 1 and 3, and head group cresting into the tunnel 2 space. Tunnels 1 and 3 may represent the principal entrance and exit routes for substrates and product, that place the headgroup in proximity of the active site.
