Extended Data Fig. 8. Characterization of sgTMEM164-C123Y base-edited cells.
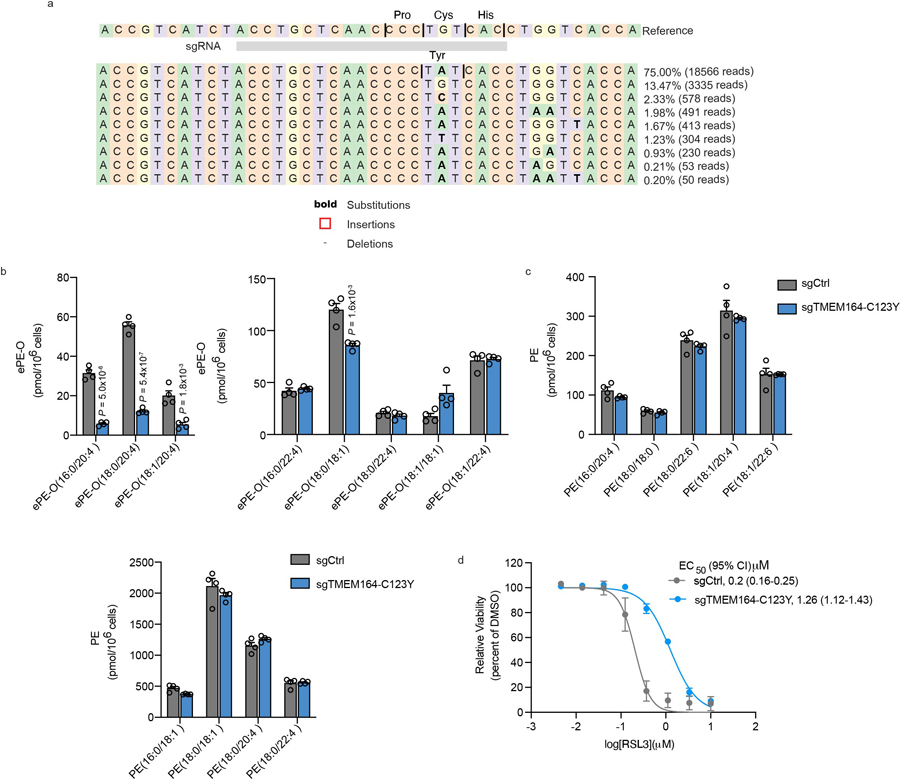
a, Confirmation of base-editing of TMEM164 genomic DNA in 786-O cells at the sgRNA target site for sgTMEM164-C123Y base-edited cell population by next-generation sequencing analysis. b, Measurement of C20:4 (left) and additional (right) ePE-O lipids in sgCtrl and sgTMEM164-C123Y base-edited cell populations. c, Measurement of diacyl PE lipids in sgCtrl and sgTMEM164-C123Y base-edited 786-O cell populations. d, Cell viability of sgCtrl and sgTMEM164-C123Y base-edited cell populations treated with the indicated concentrations of the GPX4 inhibitor RSL3 measured at 24 h post-treatment. For b and c data represent mean values ± S.E.M from four independent experiments per group. For d, data represent mean values ± S.E.M. from two independent experiments. P-values were derived using a Two-sided Student’s t-test performed relative to sgCtrl cells.
