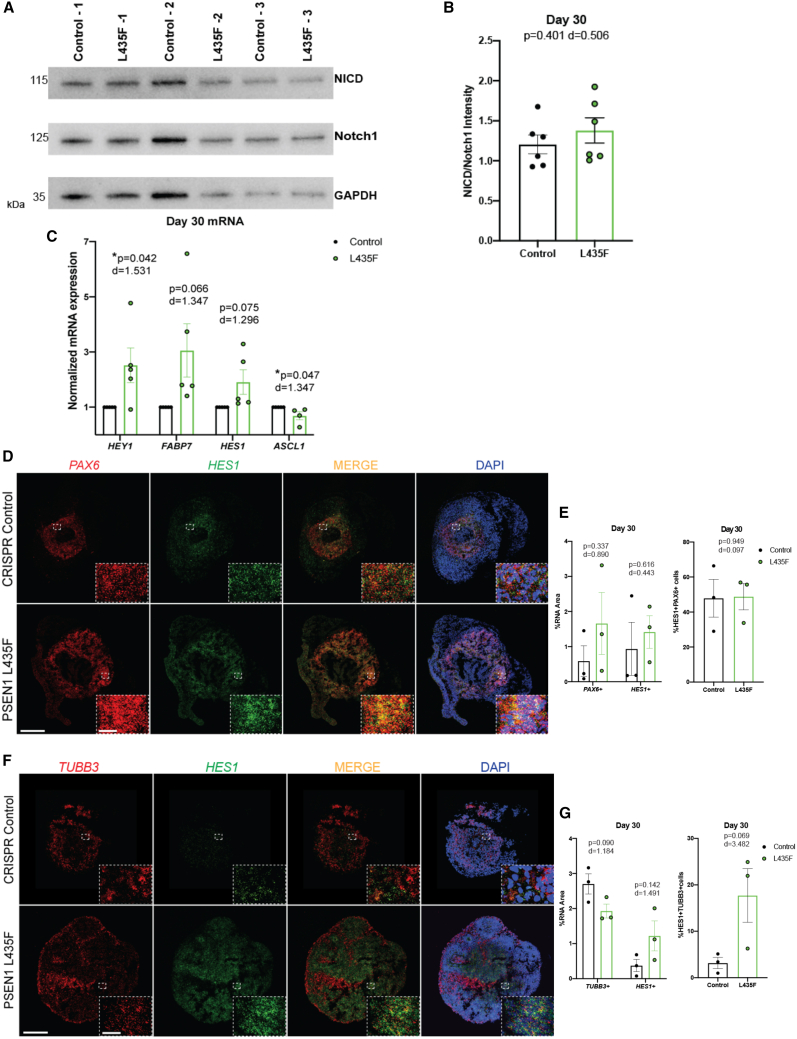
Figure 4.
PSEN1 L435F increases Notch signaling
(A) Western blot results of NICD and Notch1 in male control and PSEN1 L435F hCSs at 30 DIV. GAPDH is used as a loading control. Image depicts three independent hCS replicates on one gel.
(B) Quantification of western blot results graphed as the ratio of NICD/Notch1. N = 6 independent replicates; 50 μg protein from 4 pooled hCSs per replicate. p value result of unpaired t test and Cohen’s d effect size are displayed.
(C) qRT-PCR results of Notch target gene expression in male hCSs. N = 5 independent replicates; 500 ng of RNA from 4 pooled hCSs per experiment.
(D) RNAScope images of PAX6, HES1, and DAPI at 30 DIV in male hCSs. Scale bars, 500 and 25 μm (inset).
(E) Quantification of PAX6+ and HES1+ cells, graphed as the ratio of mRNA to DAPI expression, and the colocalization of PAX6 and HES1. N = 3 hCSs.
(F) RNAScope images of TUBB3, HES1, and DAPI at 30 DIV.
(G) Quantification of TUBB3+ and HES1+ cells, graphed as the ratio of mRNA to DAPI expression, and the colocalization of TUBB3 and HES1. N = 3 hCSs. Data are presented as mean ± SEM. p values from unpaired t tests and Cohen’s d are displayed.