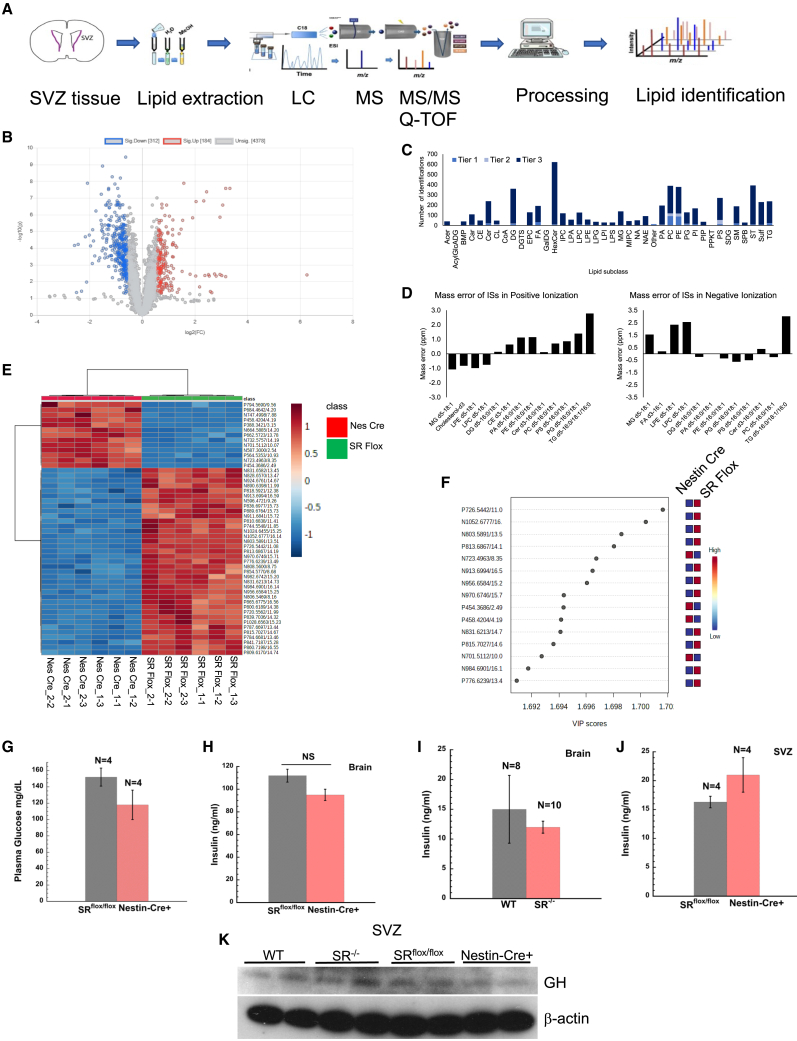
Figure 4.
Global lipidomic profile in the SVZ of nestin-cre+ mice
(A) Schematic of lipidomics workflow.
(B) Parametric volcano plot showing fold change against p value adjusted for false discovery rate. Blue circles indicate downregulation (312 lipids), red indicates upregulation (184 lipids) of lipids. Unassigned lipids are shown in gray.
(C) Lipid identification tier. In tier 1 (404 features), 2 (146 features), and 3 (remaining features).
(D) Mass error of internal standards.
(E) Heatmap of top 50 lipids ranked by p value.
(F) Plot shows partial least squares-discriminant analysis with variable importance in prediction scores of 15 most important lipids.
(G) Plasma glucose levels in SRflox/flox and nestin-cre+ mice.
(H and I) Insulin concentration (ng/mL) in mice whole brain homogenates.
(J) Insulin concentration (ng/mL) in mice SVZ homogenates. Error bars represent SD. NS = nonsignificant.
(K) Expression of growth hormone in mice SVZ. Error bars represent SD. Data are representative of three independent experiments. Tissues were pooled from n = 5–8 mice per experiment. For SVZ lipidomics, each genotype comprised tissues from 5 to 8 mice and injections in triplicate (see method for details).