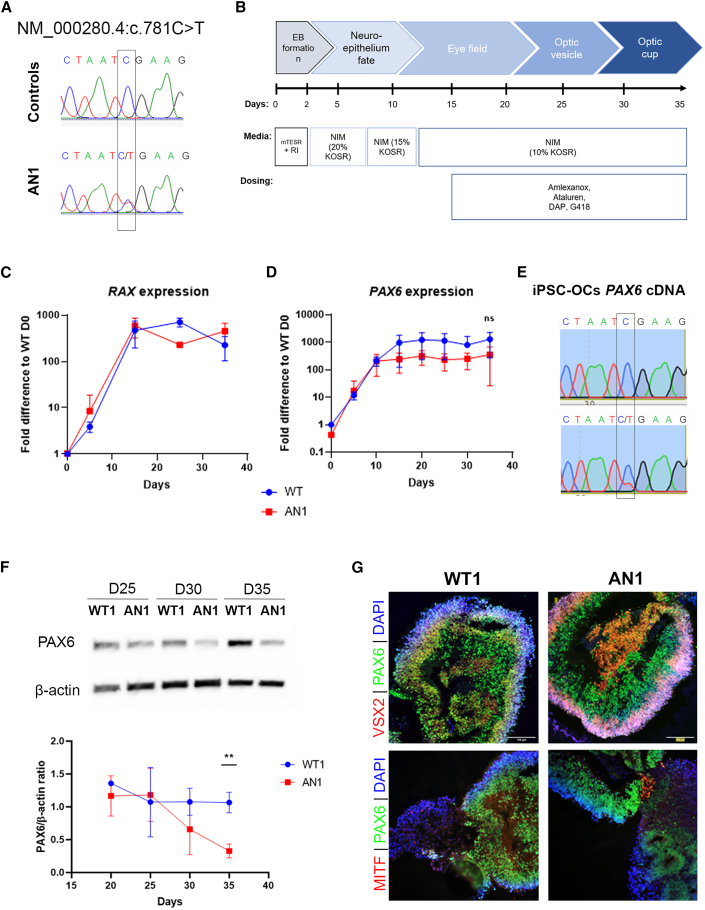
Figure 1.
Generation of AN iPSC-derived OCs
(A) Direct sequencing of PAX6 exon 10, showing the heterozygous nonsense c.781C>T change in AN1 patient iPSCs. This variant was not detected in control lines. (B) Schematic of the differentiation strategy of control WT1 and WT2 and patient-derived AN1 iPSCs into 3D OCs (35 days). Data from the WT1 control line are included as an example. Dosing experiments with TRIDs were performed from day 15 onwards. (C and D) qRT-PCR transcript analysis of the EFTFs RAX and PAX6 during 35 days of differentiation in control (WT1, blue) and aniridia (AN1, red) iPSCs. Values were normalized to day 0 and to the internal housekeeping gene GAPDH. Data represents means and SD of 3 biological replicates. (E) RT-PCR followed by Sanger sequencing of PAX6 cDNA from day 35 WT1 and AN1 iPSC-OCs. The mutated allele c.781C>T can be seen in AN1 but not in WT1 iPSC-OCs. (F) PAX6 protein analysis detected by western blot in WT and AN1 iPSC-OCs from days 25–35 of differentiation (5-day intervals). The PAX6/β-actin ratio was normalized to WT1. Data expressed as mean ± SD from n = 3 (∗∗p < 0.01, t test analysis). (G) Immunohistochemical analysis of WT1 and AN iPSC-derived OCs, showing positive staining of PAX6 (green) as well as markers for OC domains: VSX2, indicating the neural retina (red, top panel), and MITF, indicating retinal pigmented epithelium (RPE; red, bottom panel). DAPI staining (blue) shows cell nuclei. Scale bar, 100 μm.