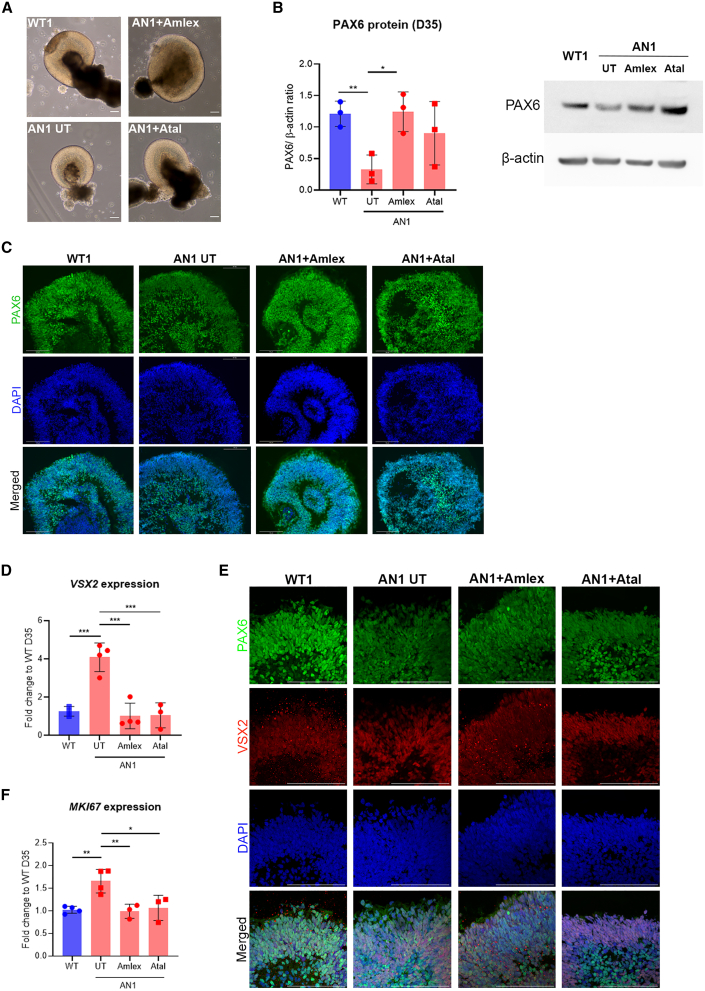
Figure 3.
Effect of translational readthrough-inducing drugs (TRIDs) in day 35 AN iPSC-OCs
(A) Bright-field images of control (WT), untreated AN (AN1 UT), amlexanox-treated AN (AN1 Amlex) and ataluren-treated AN (AN1 Atal) iPSC-OCs. Scale bar, 100 μm. (B) Quantification of PAX6 protein in treated vs. UT AN1 iPSC-OCs (red bars). The PAX6/β-actin ratio was normalized to the control (WT, blue bar). ∗p < 0.05, ∗∗p < 0.01, one-way ANOVA. Data represents means and SD of at least 3 biological replicates. (C) Immunofluorescence analysis on day 35 of differentiation, showing PAX6 staining (green) and DAPI (blue) in control (WT), AN1 UT, AN1 Amlex, and AN1 Atal iPSC-OCs. Scale bar, 100 μm. (D) qRT-PCR transcript analysis of the neural retina marker VSX2 in WT (blue bar) and AN1 UT, AN1 Amlex, and AN1 Atal samples (red bars). ∗p < 0.05, ∗∗p < 0.01, one-way ANOVA. (E) Immunohistochemical analysis showing VSX2 staining in WT, AN1 UT, and Amlex- and Atal-treated iPSC-OCs. DAPI staining is shown in blue. Scale bar, 100 μm. Red bright spots visible in WT and AN1 Amlex are background staining. (F) qRT-PCR transcript analysis of the proliferation marker MKi67 in WT (blue bar) and AN1 UT, AN1 Amlex, and AN1 Atal samples (red bars). ∗∗∗p < 0.001, one-way ANOVA. (D and F) Values were normalized to the WT and to the internal housekeeping gene GAPDH. Data represents means and SD of at least 3 biological replicates.