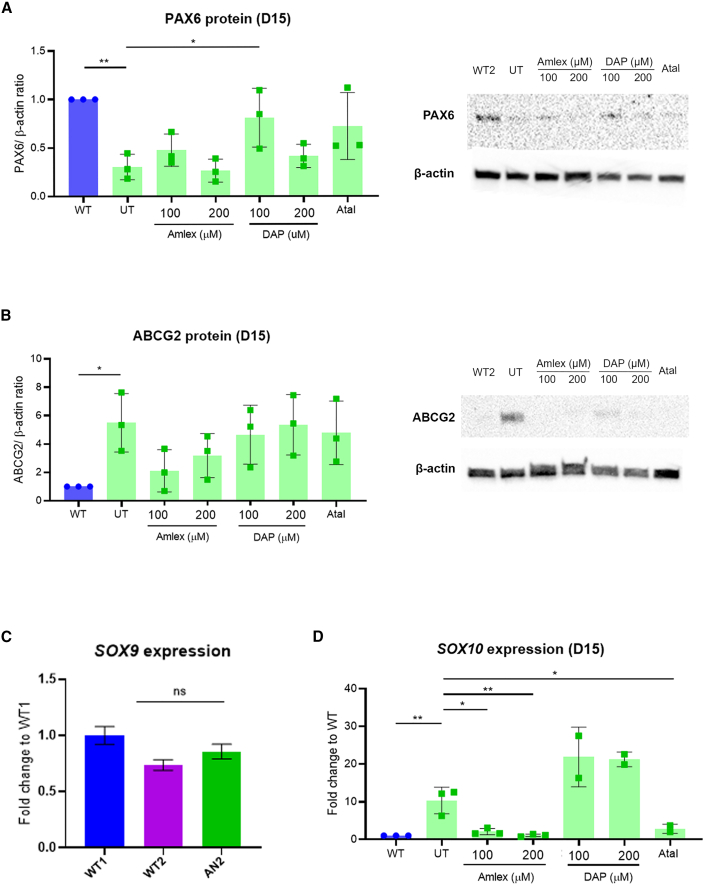
Figure 5.
TRIDs show trend improvement in PAX6 expression in AN2 iPSC-derived LESCs
(A) Quantification of PAX6 protein in UT AN2 iPSC-LESCs versus Amlex-, DAP-, and Atal-treated AN2 iPSC-LESCs (green bars). The PAX6/β-actin ratio was normalized to the control (WT, blue bar). Values on the x axis refer to compound concentrations (in μM). ∗p < 0.05, ∗∗p < 0.01, one-way ANOVA). Data represents means and SD of at least n = 3 biological replicates. (B) Quantification of ABCG2 protein detected by western blot in WT (blue bar), AN2 UT, and Amlex-, DAP-, and Atal-treated AN2 iPSC-LESCs (green bars). Values on the x axis refer to compound concentrations (in μM). The PAX6/β-actin ratio was normalized to the control (WT). Data represent means and SD of 3 biological replicates. ∗p < 0.05, one-way ANOVA. (C) Relative expression of SOX9 transcripts in WT1, WT2, and AN2 iPSC-LESCs. Significance was calculated using multiple t tests between AN and both WT lines. (D) qRT-PCR transcript analysis of the neural crest marker SOX10 in WT (blue bar) and AN UT, AN Amlex, and AN Atal samples (green bars). ∗∗∗p < 0.001, ∗∗p < 0.01, one-way ANOVA. (C and D) Values were normalized to the WT and to the internal housekeeping gene GAPDH. Data represents means and SD of at least 3 biological replicates.