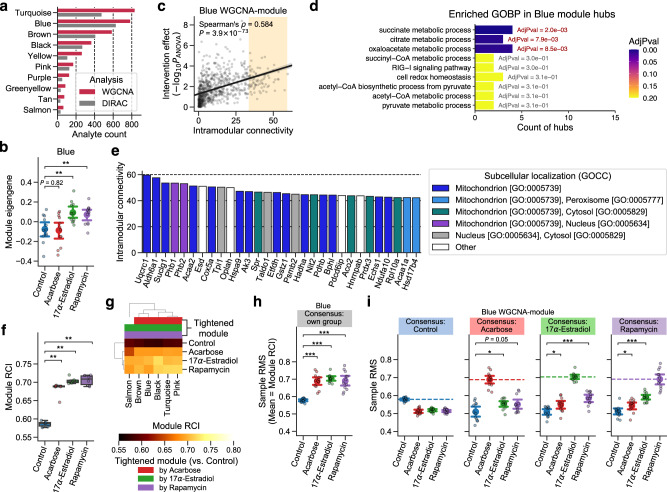
Fig. 3. Lifespan-extending interventions increased proteomic profile conservation in data-driven modules.
a–e Weighted Gene Co-expression Network Analysis (WGCNA) of the LC-M001 proteomics data (see Supplementary Data 2 and 3 for complete results). a The number of proteins in each WGCNA-identified module. WGCNA: proteins used in WGCNA, DIRAC: proteins retained after the processing for Differential Rank Conservation (DIRAC) analysis f–i. b Distributions of sample’s module eigengene values for the Blue module. Data: the mean (dot) with 95% confidence interval (CI) (bar); n = 12 mice. **P < 0.01 by two-sided Welch’s t-tests after the Benjamini–Hochberg adjustment across three comparisons. c Relationship between the intervention effect on each protein and their respective intramodular connectivity in the Blue module. The P-value of y-axis corresponds to the main effect of intervention on each protein level by Analysis of Variance (ANOVA). The line is the ordinary least squares (OLS) linear regression line with 95% CI, and the orange-colored background reflects the range of top 10% hub proteins (79 proteins). n = 787 proteins. d Enriched Gene Ontology Biological Process (GOBP) terms in the top 10% hub proteins of the Blue module. Significance was assessed using overrepresentation tests after the Benjamini–Hochberg adjustment across 81 terms. Only the GOBP terms that exhibited nominal P < 0.05 are presented. AdjPval: adjusted P-value from the overrepresentation test. e Top 30 hub proteins of the Blue module. GOCC: GO Cellular Component. f–i DIRAC analysis of the LC-M001 proteomics data using WGCNA-identified modules (see Supplementary Data 4 for complete results). f, g Overall distribution of module rank conservation index (RCI). Data in f: median (center line), [Q1, Q3] (box limits), [xmin, xmax] (whiskers), where Q1 and Q3 are the 1st and 3rd quartile values, and xmin and xmax are the minimum and maximum values in [Q1 − 1.5 × IQR, Q3 + 1.5 × IQR] (IQR: the interquartile range, Q3 − Q1), respectively; n = 6 modules. **P < 0.01 by two-sided Mann–Whitney U-tests after the Benjamini–Hochberg adjustment across three comparisons. The top color columns in g highlight the modules that exhibited (1) the significant intervention effect on module RCI (ANOVA after with the Benjamini–Hochberg adjustment across six modules) and (2) significantly higher RCI in intervention group than control group (i.e., tightened module; the post hoc two-sided Welch’s t-tests after the Benjamini–Hochberg adjustment across three comparisons). h, i Sample rank matching score (RMS) distributions for the Blue module. Dashed line in i indicates the mean of RMSs for the rank consensus group (i.e., RCI). Data: the mean (dot) with 95% CI (bar); n = 12 mice. *P < 0.05, ***P < 0.001 by two-sided Welch’s t-tests after the Benjamini–Hochberg adjustment across three (h) or six (two comparisons × three rank consensus; i) comparisons.