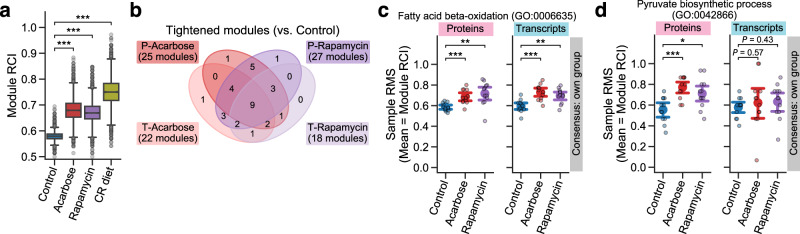
Fig. 4. Lifespan-extending interventions increased the conservation of proteomic modules through transcriptional or post-transcriptional alteration.
a Differential Rank Conservation (DIRAC) analysis of the M001-related transcriptomics data using Gene Ontology Biological Process (GOBP)-defined modules (see Supplementary Data 5 for complete results). Presented is the overall distribution of module rank conservation index (RCI). CR: calorie restriction. Data: median (center line), 95% confidence interval (CI) around median (notch), [Q1, Q3] (box limits), [xmin, xmax] (whiskers), where Q1 and Q3 are the 1st and 3rd quartile values, and xmin and xmax are the minimum and maximum values in [Q1 − 1.5 × IQR, Q3 + 1.5 × IQR] (IQR: the interquartile range, Q3 − Q1), respectively; n = 3912 modules. ***P < 0.001 by two-sided Mann–Whitney U-tests after the Benjamini–Hochberg adjustment across three comparisons. b–d DIRAC comparison analysis between the LC-M001 proteomics and M001-related transcriptomics data using GOBP-defined modules (see Supplementary Data 6 for complete results). b Venn diagram of the modules that exhibited (1) the significant intervention effect on module RCI (Analysis of Variance (ANOVA) after the Benjamini–Hochberg adjustment across 312 (156 modules × two omics datasets) tests) and (2) significantly higher RCI in intervention group than control group (i.e., tightened module; the post hoc two-sided Welch’s t-tests after the Benjamini–Hochberg adjustment across four (two comparisons × two omics datasets) comparisons). P: proteomics, T: transcriptomics. c, d Sample rank matching score (RMS) distributions for an example of the tightened modules in both proteins and transcripts (c; GO:0006635, fatty acid β-oxidation) or specifically in proteins (d; GO:0042866, pyruvate biosynthetic process). Data: the mean (dot) with 95% CI (bar); n = 12 mice. *P < 0.05, **P < 0.01, ***P < 0.001 by two-sided Welch’s t-tests after the Benjamini–Hochberg adjustment across four (two comparisons × two omics datasets) comparisons.