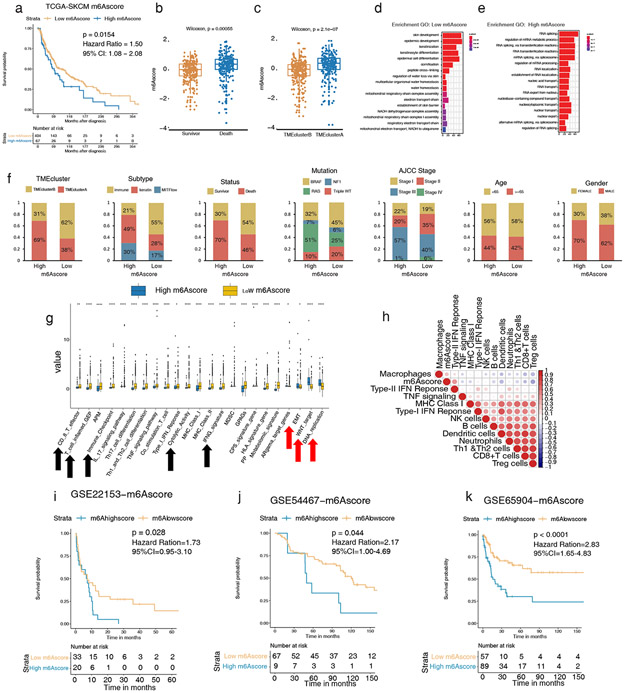
Figure 4: High m6A modifications correlate to worse tumor phenotypes and survival in melanoma validation cohorts.
(a) Survival analyses based on high and low m6Ascores of 473 patients in TCGA-SKCM show low m6Ascore has a significant survival advantage (HR, 1.50; 95% CI, 1.08–2.08; p = 0.0154). (b–c) Differences of m6Ascore and survival status among TMEclusters in TCGA-SKCM (p = 2.1e-07 and p = 5.5e-4 respectively, Wilcoxon test). (d–e) GO analysis of DEG presented higher skin cell differentiation and sufficient oxygen reaction in low m6Ascore (d) and higher active RNA modification and transcription in high m6Ascore (e) in TCGA-SKCM. (f) The clinicopathological characteristics in low m6Ascore are associated with more TMEclusterB, more immune subtype, better survival status, more triple WT, and earlier AJCC stage. Age and gender were allocated evenly. (g) Relative gene signature intensities enrich distinct categories in high and low m6Ascore: immune signal activation for low m6Ascore and cell active proliferation, transformation, and DNA mismatch for high m6Ascore. (h) Correlation of m6Ascore and infiltrating immune cells and immune response by Spearman correlation analysis. m6Ascore is negatively correlated with active immune singling in the TCGA-SKCM cohort. (i–k) Survival analyses based on high and low m6Ascore in GSE22153 (HR, 1.73; 95% CI, 0.95–3.10; p = 0.028), in GSE54456 (HR, 2.17; 95% CI, 1.00–4.69; p = 0.044), and in GSE65904 (HR, 2.83; 95% CI, 1.65–4.83; p < 0.0001), indicating that low m6Ascore has a significant survival advantage.