Figure 1.
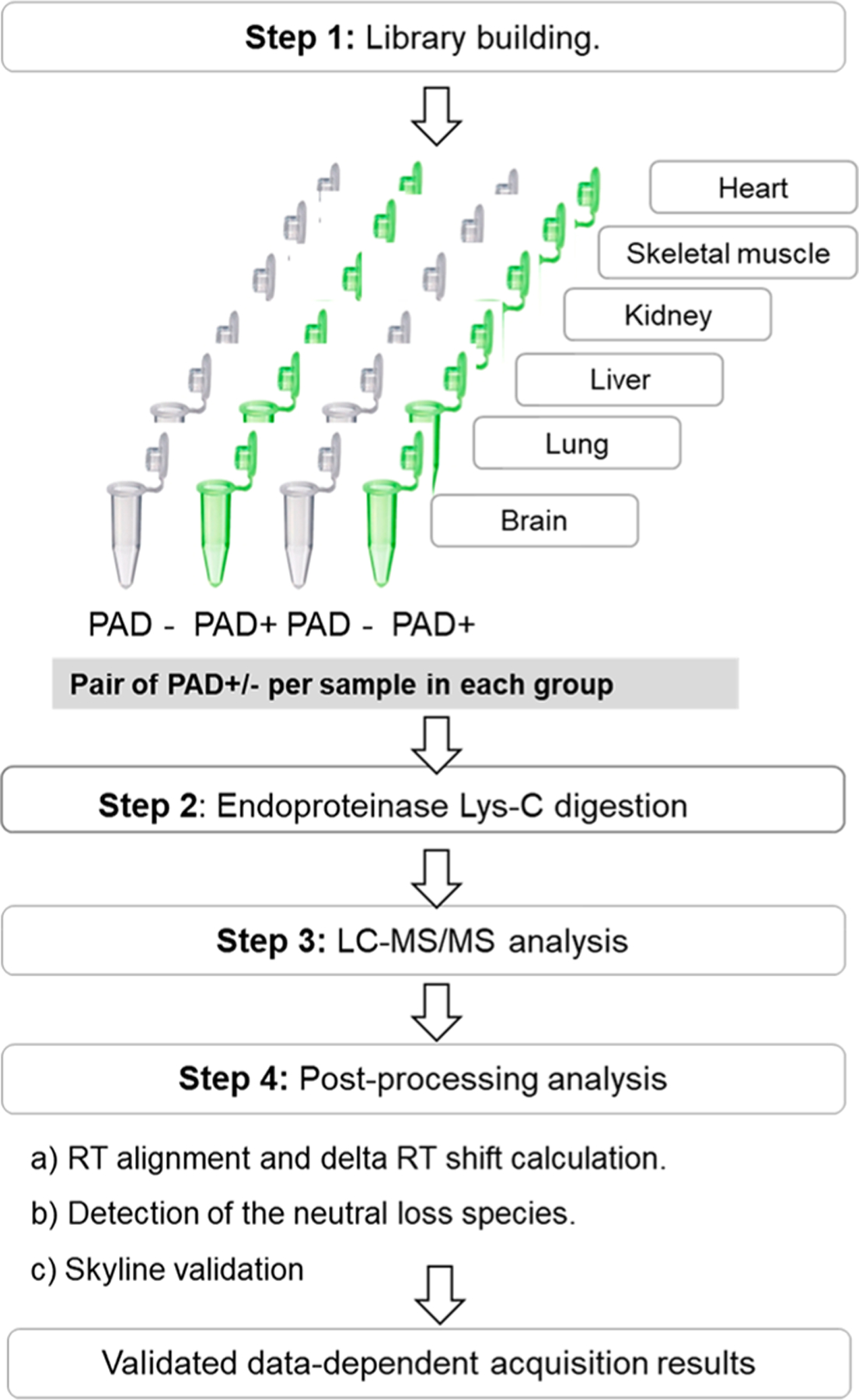
Flowchart outlining the workflow for the hyper-citrullinated library generation pipeline. A total of six tissues were isolated from three male wild-type mice. The tissues were snap-frozen, homogenized, and solubilized, and the total protein concentration was determined using a BCA protein assay kit. Step 1. Library building. In brief, each sample was split into two samples; one was treated with PAD cocktail (five isoforms, ratio 1:20) to cover all potential citrullinated targets. The corresponding second sample was treated with water under the same condition. Step 2. Digestion with LysC (ratio 1:20), followed by LC–MS/MS (TripleTOF 5600). Step 3. Postprocessing analysis with algorithm that examines the MS runs for the evidence of citrullinated site and Skyline validation.
