Figure 1.
Chromosome-level assembly and annotation of Ae. speltoides.
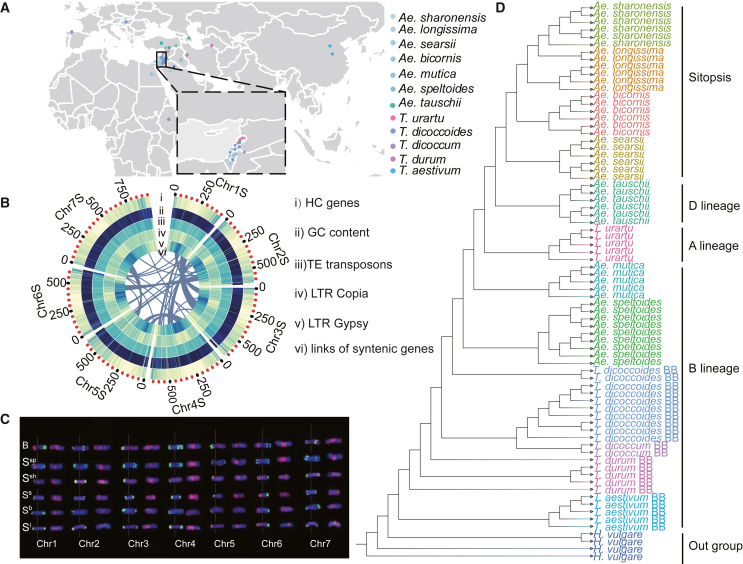
(A) Geographic distribution of the 12 species used in this research: Ae. bicornis, Ae. longissima, Ae. mutica, Ae. searsii, Ae. sharonensis, Ae. speltoides, Ae. tauschii, T. aestivum, T. dicoccoides, T. dicoccum, T. durum, and T. urartu.
(B) Circos plot of genomic features of the assembled Ae. speltoides genome: (i) density of HC genes, (ii) distribution of GC content, (iii) TE density along each chromosome, (iv) density of Copia TEs, (v) density of Gypsy elements, and (vi) links between syntenic genes.
(C) FISH results with the probes (left) Oligo-pSc119.2 (green) + Oligo-pTa71 (red) and the probe (right) Oligo-(GAA)7 (red) for the chromosomes of Ae. speltoides (Ssp), Ae. searsii (Ss), Ae. bicornis (Sb), Ae. longissima (Sl), Ae. sharonensis (Ssh), and the wheat B subgenome (B). The numbers 1–7 represent homoeologous chromosome groups 1–7, respectively.
(D) Phylogenetic tree constructed with data from 13 species using the neighbor-joining method; barley was used as the outgroup species.