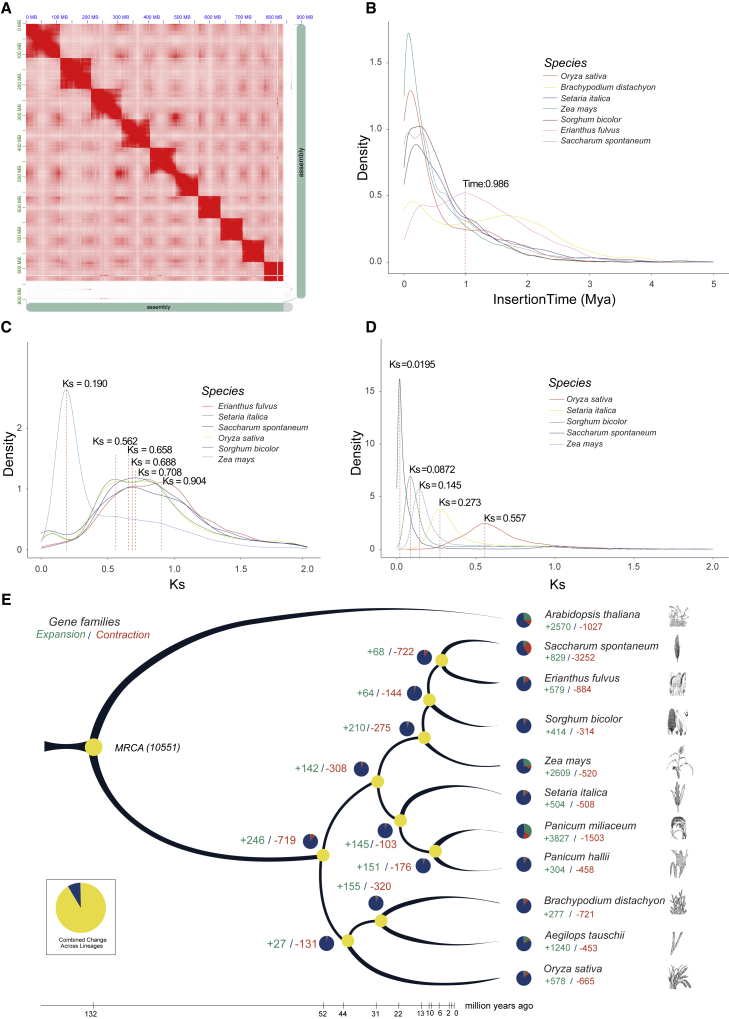
Figure 2.
Evolutionary characteristics of the E. fulvus genome.
(A) Hi-C interaction map.
(B) Depiction of LTR evolution.
(C) Representation of whole-genome duplication (WGD) events during E. fulvus evolution based on synonymous substitution rates (Ks).
(D) Density distribution of Ks values for paired genes calculated within each syntenic block between E. fulvus and other species.
(E) Genome evolution based on single-copy orthologs of 10 species from the PACMAD and BOP clades. A. thaliana was used as an outgroup. Numbers in green indicate expanded gene families, and numbers in red indicate contracted gene families.