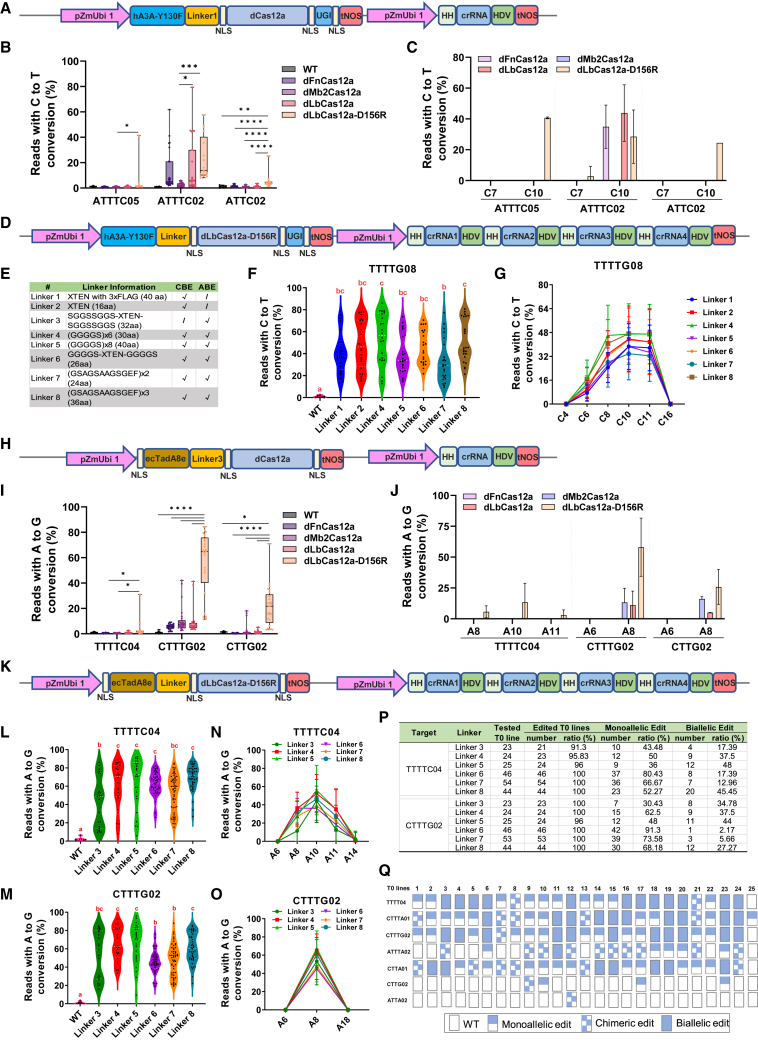
Figure 1.
Development of CRISPR–Cas12a–based cytosine base editors (CBEs) and adenine base editors (ABEs) in rice.
(A) Schematic of the dual RNA polymerase II (Pol II) promoter system for dCas12a-based CBE and crRNA expression. pZmUbi1, maize ubiquitin promoter; NLS, nuclear localization signal; UGI, uracil DNA glycosylase; tNOS, nopaline synthase terminator; HH, hammerhead ribozyme; HDV, hepatitis delta virus ribozyme.
(B) Assessment of the C–to–T editing efficiency of four variants of dCas12a-based CBEs at sites ATTTC05, ATTTC02, and ATTC02 in transgenic rice plants.
(C) Editing window of four dCas12a-CBEs in transgenic rice plants at sites ATTTC05, ATTTC02 and ATTC02.
(D) Schematic of the dual Pol II promoter–based and multiplexed dLbCas12a-D156R CBE editing system.
(E) The list of linkers used between deaminases and dLbCas12a–D156R.
(F) Assessment of seven dLbCas12a–D156R CBEs with different linkers in regenerated rice calli at the TTTTG08 site. Each dot represents an independent callus. The first quartile, median, and third quartile are shown as dotted lines.
(G) Editing windows of seven dLbCas12a–D156R CBEs with different linkers in regenerated rice calli at the TTTTG08 site.
(H) Schematic of the dual Pol II promoter system for dCas12a–based ABE and crRNA expression.
(I) Assessment of A–to–G editing efficiency of four variants of dCas12a-based ABEs at sites TTTTC04, CTTTG02, and CTTG02 in transgenic rice plants.
(J) Editing window of four dCas12a– ABEs in transgenic rice plants at sites TTTTC04, CTTTG02, and CTTG02.
(K) Schematic of the dual Pol II promoter–based multiplexed dLbCas12a-D156R ABE editing system.
(L and M) Assessment of six dLbCas12a–D156R ABE editors with different linkers in transgenic rice plants at TTTTC04 and CTTTG02 sites. The first quartile, median, and third quartile are shown as dotted lines. Each dot represents an independent line.
(N and O) Editing windows of six dLbCas12a-D156R ABE editors with different linkers in transgenic rice plants at TTTTC04 and CTTTG02 sites.
(P) A–to–G base editing frequency of dLbCas12a–D156R–based ABEs at TTTTC04 and CTTTG02 sites.
(Q) Genotypes of 25 T0 lines edited by the dLbCas12a–D156R–linker 5–ABE at seven target sites. Wild type, chimeric edits, monoallelic edits, and biallelic edits are denoted as empty rectangles, dotted rectangles, half-filled rectangles, and fully filled rectangles, respectively.
To define genotypes in (P) and (Q): T0 lines with an A–to–G mutation frequency lower than 10% were regarded as wild type; T0 lines with an A–to–G mutation frequency from 10% to 30% were regarded as having chimeric edits; T0 lines with an A–to–G mutation frequency from 30% to 75% were regarded as having monoallelic edits; and T0 lines with an A–to–G mutation frequency higher than 75% were regarded as having biallelic edits. NGS of PCR amplicons was used to detect mutations in regenerated rice calli or T0 lines. All NGS data were analyzed with CRISPR RGEN tools. Data in (C), (G), (J), (N), and (O) are presented as mean values ± standard deviation. p values in (B) and (I) were obtained using a two-tailed Student’s t-test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Different letters indicate significant differences (p < 0.05; one-way ANOVA, Tukey’s test) in (F), (L), and (M).