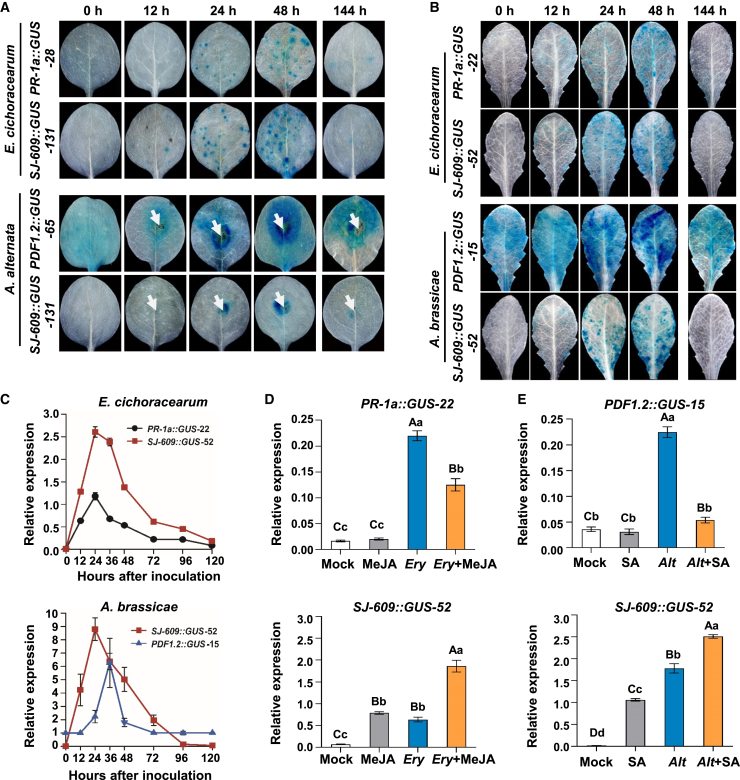
Figure 4.
Induction of GUS activity in transgenic tobacco and Arabidopsis upon infection with biotrophic and necrotrophic pathogens.
(A) GUS staining of leaves from representative PR-1a::GUS-28, SJ-609::GUS-131, and PDF1.2::GUS-65 transgenic tobacco lines infected with the biotrophic pathogen E. cichoracearum or the necrotrophic pathogen A. alternata. White arrows indicate the site of A. alternata inoculation. To withdraw the pathogen challenge, fungicide (fenaminosulf) was sprayed onto infected leaves 72 h after inoculation. GUS staining was performed 72 h after withdrawal of the challenge. Ten transgenic tobacco lines per construct were used for analysis of GUS activity.
(B) GUS staining of leaves from representative PR-1a::GUS-22, SJ-609::GUS-52, and PDF1.2::GUS-15 transgenic Arabidopsis lines infected with the biotrophic pathogen E. cichoracearum or the necrotrophic pathogen Alternaria brassicae. Five transgenic Arabidopsis lines per construct were used for analysis of GUS activity.
(C)GUS expression patterns of representative PR-1a::GUS-22, PDF1.2::GUS-15, and SJ-609::GUS-52 transgenic Arabidopsis lines induced by E. cichoracearum (above) and A. alternata (below). Values are the means of three technical replicates with SD. Similar results were obtained for three repeated experiments. E. cichoracearum and A. alternata were sprayed as 107 spore/ml spore suspensions.
(D and E)GUS expression in PR-1a::GUS-22, PDF1.2::GUS-15, and SJ-609::GUS-52 transgenic Arabidopsis lines with the indicated treatments. Mock, deionized water as a mock control for SA treatment (E) or 0.1% ethyl alcohol as a mock control for MeJA treatment (D); MeJA, sprayed with 0.5 mM MeJA; Ery, inoculated with E. cichoracearum; Ery + MeJA, inoculated with E. cichoracearum, then sprayed with 0.5 mM MeJA 24 h later; SA, sprayed with 1 mM SA; Alt, inoculated with A. alternata; Alt + SA, inoculated with A. alternata, then sprayed with 1 mM SA 24 h later. Results in (D and E) are averages of three biological replicates with SD. Lowercase and capital letters above bars indicate significant differences according to Duncan’s multiple comparison procedure at the 0.05 (P < 0.05) or 0.01 level (P < 0.01), respectively. GUS expression levels in (C), (D), and (E) were quantified by qRT-PCR of cDNA prepared from RNA extracted from treated or infected leaves. AtACT2 served as the reference gene. GUS and AtACT2 primers for qRT-PCR are shown in Supplemental Table 9.