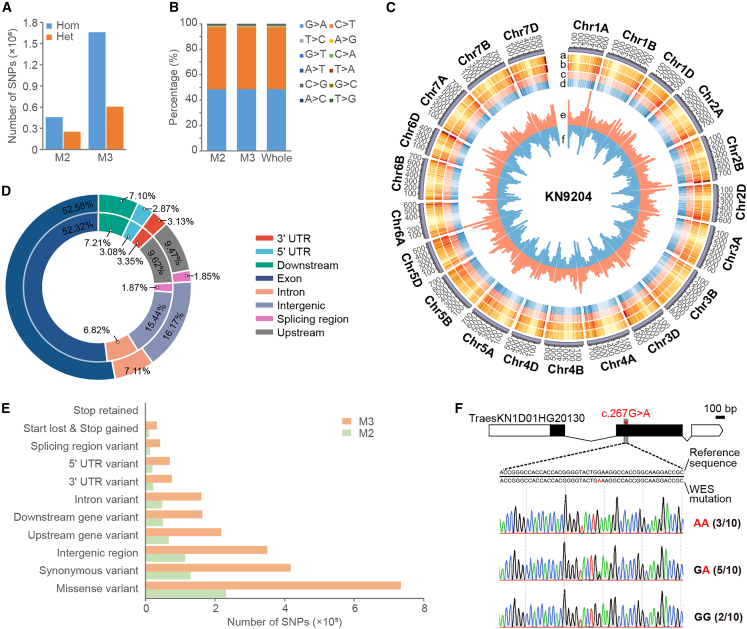
Figure 3.
Characterization of mutations in the KN9204 EMS-mutagenized library.
(A) The numbers of homozygous and heterozygous single-nucleotide polymorphism (SNP) mutations in the M2 and M3 populations. Hom, homozygous; Het, heterozygous.
(B) The proportions of nucleotide mutations in M2, M3, and the whole mutant population. EMS-type mutations (C → T and G → A) accounted for more than 97% of the SNPs.
(C) Circos plot of important mutation indicators in the EMS-mutagenized population. From the outer to the inner circle: (a) probe density, (b) mutation density in exons, (c) density of heterozygous mutations (light red), (d) density of homozygous mutations (light blue), (e) numbers of severe mutations (orange), and (f) numbers of SNPs on the chromosome (light blue).
(D) The distribution of SNPs in gene structures, with mutations in the M2 population plotted on the inner circle and those in M3 on the outer one. More than half of the SNPs were located in exons in both populations.
(E) Functional annotation of SNP-caused mutations in genes. Mutations in the M2 population are marked in green and those in M3 in orange.
(F) Validation of WES-identified EMS mutations via Sanger sequencing. Numbers indicate the frequency of specific nucleotides from multiple sequencing results.