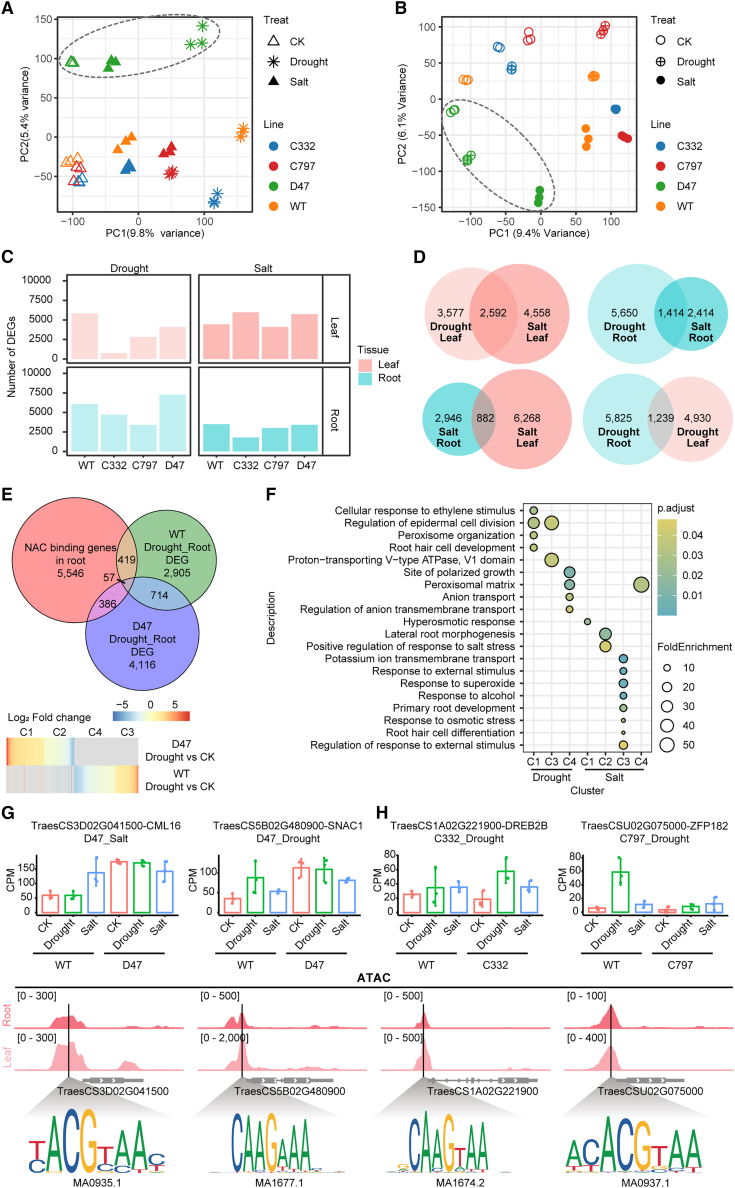
Figure 7.
Transcriptional regulation of NAC TF targets in response to drought and salt stresses.
(A and B) Principal component analysis (PCA) of transcriptome data for root samples (A) and leaf samples (B) under different conditions. Each symbol represents one sample. As indicated, samples of different mutant lines, tissues, and treatments are displayed in different shapes and colors. Dashed ovals separate the samples of the D47 line from others.
(C) Numbers of DEGs in the roots and leaves of each mutant line under different conditions.
(D) Venn diagrams show the overlap of salt or drought stress–responsive DEGs in different tissues of the mutant line C332 compared with the control.
(E) Schematic of the strategy used to identify target candidates of TraesCS5A02G242600 (D47). The KN9204 (WT) and D47 mutant line-specific drought-responsive genes and genes with different response levels (log2(FCKN9204/FCmutant) >1) are summed as TraesCS5A02G242600 (D47)-affected drought-responsive genes. Among them, genes with NAC TF binding sites in the open promoter region are considered to be targets of NAC TFs during drought treatment. The heatmap shows the mRNA fold change of 862 identified drought-responsive TraesCS5A02G242600 target candidates in the roots of the WT or the D47 mutant line.
(F) Enriched GO terms in the identified TraesCS5A02G242600 (D47) target candidates under drought or salt stress.
(G and H) Expression patterns of the NAC TF target candidates TaSNAC1-D, OsCML16(G), OsDREB-2B, and OsZFP182(H) under CK, drought, or salt-stress conditions in the WT and various mutant lines (top). These candidates harbor different NAC TF binding motifs under the ATAC-seq peak in their promoter regions in root or leaf tissue, as indicated (bottom).