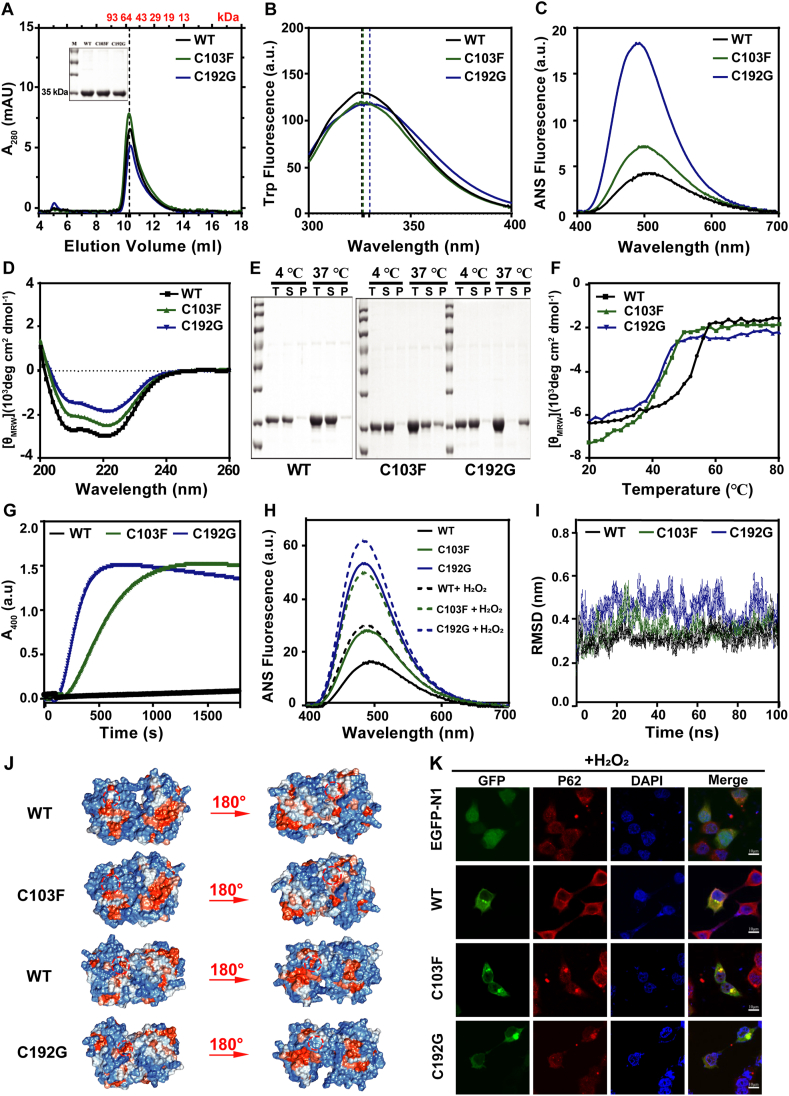
Figure 1.
The pathogenic cysteine mutants (C103F and C192G) disrupted the structure of PMM2. (A) SEC profiles of 100 μL 10 μM purified PMM2-WT, C103F, and C192G proteins. The inset shows the SDS-PAGE analysis of the purified proteins. (B) Intrinsic Trp fluorescence spectra, (C) ANS fluorescence spectra, and (D) Far-UV CD spectra of the 10 μM WT, C103F, and C192G. (E) SDS-PAGE analysis of the soluble and insoluble fractions of protein solutions incubated at 37 °C for 4 h, T: total, S: supernatant, P: precipitate. (F) The ellipticity at 222 nm of Far-UV CD upon temperature-series changes. (G) Thermal aggregation kinetics of the proteins obtained by heating the protein solutions at 50 °C. (H) ANS fluorescence spectra of the 10 μM WT, C103F, and C192G after 1 mM H2O2 treatment for 12 h at 4 °C. (I) Cα root mean square deviation (RMSD) of the dimeric WT and Cys mutants from the initial structure time for all simulations. (J) Surface electrostatic potentials of the dimeric WT and Cys mutants. The Cys mutants altered the distribution of charged residues and the representative areas were indicated by dotted circles. (K) Representative confocal images of HEK293T cells with exogenous expression of the WT, C103F, and C192G fused by GFP at the N-terminus with 2 mM H2O2 treatment. Exogenously expressed PMM2 proteins were visualized by GFP (green), the aggresomes were marked by P62 (red), and the nucleus was stained by DAPI (blue). Scale bars = 10 μm.