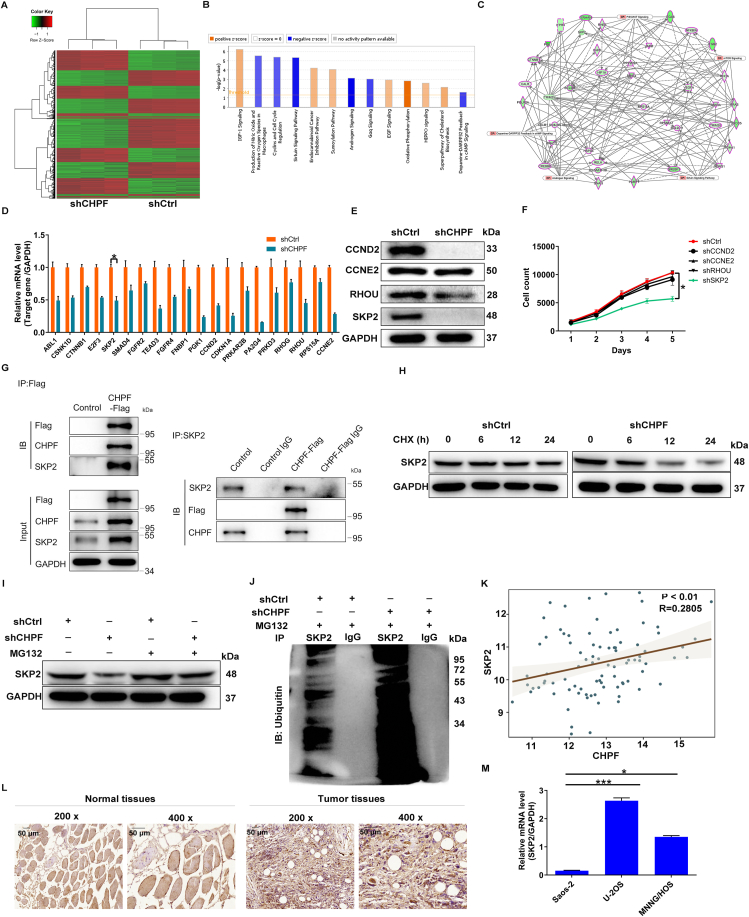
Figure 4.
SKP2 was identified as the downstream target of CHPF regulating osteosarcoma. (A) PrimeView human gene expression array (3 v 3) was performed to identify differentially expressed genes in MNNG/HOS cells with or without CHPF knockdown. (B) The enrichment of the DEGs in canonical signaling pathways was analyzed by IPA. (C) The IPA analysis was performed to produce the CHPF-related interaction network. (D, E) The expression of several most significantly down-regulated DEGs was further determined via qRT-PCR (D) and Western blotting (E). (F) Celigo cell counting assay was performed to detect the inhibition of cell proliferation of MNNG/HOS cells after infecting shCCND2, shCCNE2, shRHOU or shSKP2 corresponding lentiviruses. (G) Co-IP assay was used to verify whether there was protein interaction between CHPF and SKP2. (H) The level of SKP2 protein in MNNG/HOS cells with CHPF depletion was detected following 50 μg/mL CHX treatment for indicated times. (I) After MG-132 treatment, the level of SKP2 protein in MNNG/HOS cells with CHPF depletion was examined. (J) The lysate of MNNG/HOS cells with CHPF depletion was immunoprecipitated using SKP2 and IgG antibodies, and the Western blotting was performed to examine the ubiquitination of SKP2. (K) There was a positive correlation between CHPF level and SKP2 expression through analyzing the RNA-seq data collected from TARGET database. (L, M) The protein level of SKP2 in osteosarcoma/normal tissues and the mRNA level of SKP2 in osteosarcoma cell lines were detected via IHC staining (L) and qRT-PCR (M), respectively.