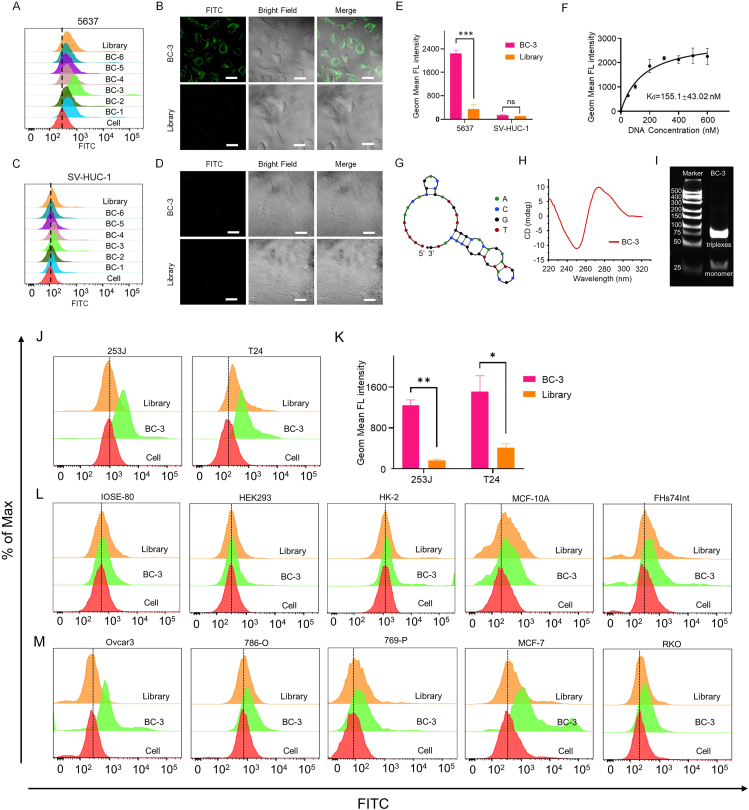
Figure 2.
Characterization and specific binding of aptamer BC-3. 5637 cells (A) and SV-HUC-1 cells (C) were incubated with FITC-labeled BC-1 to BC-6 at 4 °C, with Library or cells only used as a control. The binding ability was analyzed by flow cytometry. The FITC-labeled BC-3 (250 nM) binding to 5637 cells (B) and SV-HUC-1 (D) was displayed by confocal microscopy, with Library used as a control. Images were captured by confocal microscopy. (E) Statistical evaluation of flow cytometry results (∗∗∗P < 0.005; ns, not significant). (F) The dissociation constant of BC-3 for 5637 cells was analyzed by flow cytometry. (G) Predicted secondary structure of aptamer BC-3. (H) CD spectra of BC-3. (I) 12% PAGE gel electrophoresis analysis of BC-3 (scale bar = 40 nm). (J) Affinity of BC-3 to cell lines 253J and T24 was analyzed by flow cytometry. (K) Statistical evaluation of flow cytometry results (Comparison: BC-3 vs. Library: ∗P < 0.05, ∗∗P < 0.01). (L) BC-3 binding to various normal cell lines was analyzed by flow cytometry. (M) BC-3 cells recognizing with the corresponding cancer cell lines were analyzed by flow cytometry.