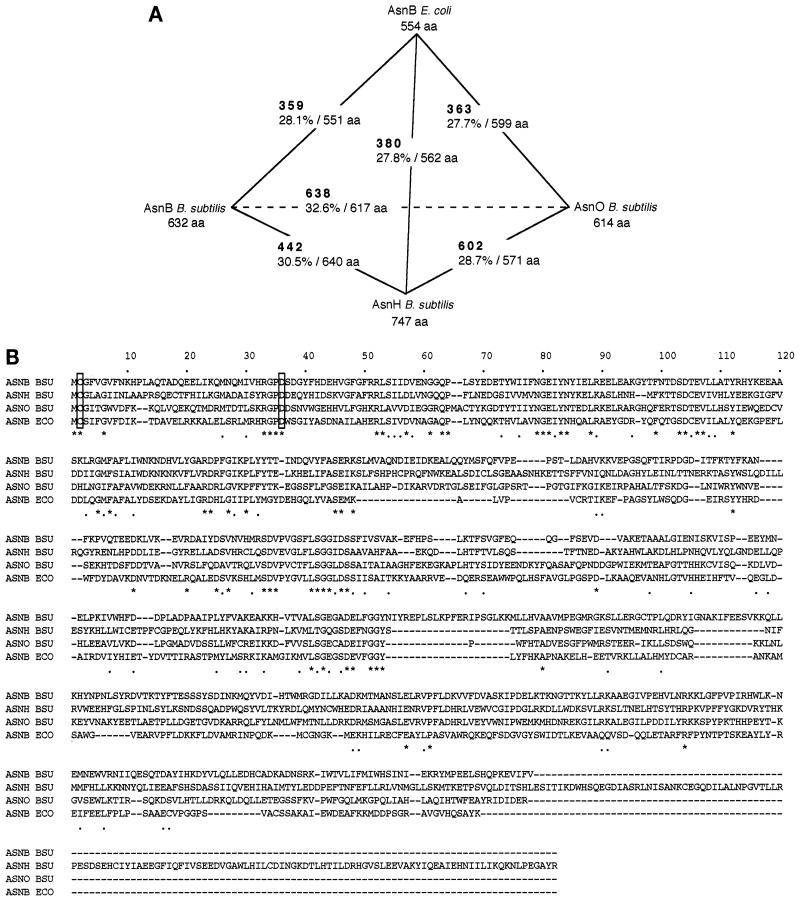
FIG. 2.
Similarity among the putative products of asnB, asnH, and asnO of B. subtilis and asnB of E. coli. (A) Similarity among the four gene products. Similarity was calculated by using the FASTA program (14) for each pairing among the four gene products. The FASTA optimized score (boldface) and sequence identity (percentage of overlapping amino acid residues) are shown. The size of each of the gene products is given as amino acid residues (aa). (B) Alignment of the amino acid sequences of the gene products. The amino acid sequence alignment of AsnB, AsnH, and AsnO of B. subtilis (ASNB BSU, ASNH BSU, and ASNO BSU, respectively) and AsnB of E. coli (ASNB ECO) was performed with the CLUSTAL W program (6). Conserved and related amino acid residues are marked with asterisks and dots beneath the sequences, respectively. Gaps introduced within the sequences to optimize the alignment are shown by hyphens. Conserved Cys and Asp residues are boxed (see text).