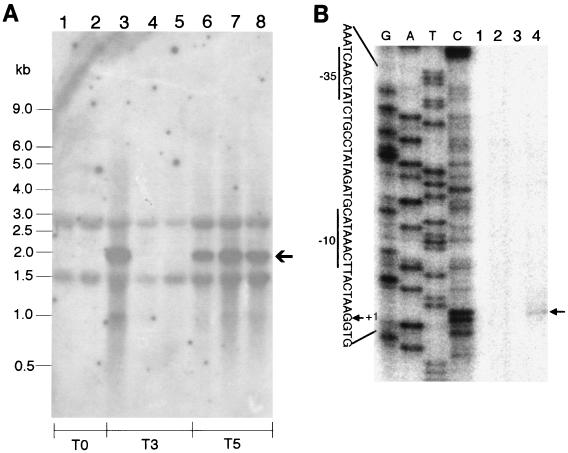
FIG. 7.
Expression of asnO depends on sigma-E. (A) Northern analysis of asnO transcription in sporulating cells of B. subtilis strains. The same specific asnO probe as for the previous experiments (Figs. 3B and 6) was used. RNAs were prepared from cells of B. subtilis strains grown in DSM. The RNA samples were taken from cells of strains 168 and ASK201 (without sigma-H) at the beginning of sporulation (T0) (lanes 1 and 2, respectively); strains 168, ASK203 (without sigma-E), and ASK202 (without sigma-F) at 3 h after the beginning of sporulation (T3) (lanes 3, 4, and 5, respectively); and strains 168, ASK205 (without sigma-K), and ASK204 (without sigma-G) at 5 h after the beginning of sporulation (T5) (lanes 6, 7, and 8, respectively). Positions of size marker RNAs and the asnO transcript (arrow) are given on the left and right, respectively. (B) Primer extension mapping of a 5′ end of the asnO transcript. The end-labeled primer (see Materials and Methods) was hybridized to RNA samples prepared from cells of strain 168 grown in DSM and was then extended with Moloney murine leukemia virus reverse transcriptase (Gibco BRL). The RNA samples were taken in exponential growth (lane 1), at the time of transition between exponential growth and stationary phase (lane 2), and at 1 h (lane 3) and 3 h (lane 4) after the beginning of sporulation. A known DNA sequence ladder (lanes G, A, T, and C) made by dideoxy sequencing reactions with the same end-labeled primer was loaded to estimate the size of the extended product. The position of the extended product is indicated by an arrow on the right. Part of the nucleotide sequence (noncoding strand) of the asnO promoter region is shown on the left. The −10 and −35 promoter regions are underlined, and the 5′ end corresponding to the transcription start site (+1) is indicated.