Acral melanoma (AM) is a rare subtype of cutaneous melanoma linked to poor prognosis, largely due to a lacking of effective targeted therapeutic strategies. Whole-genome sequencing (WGS) data revealed that AM showed a different mutation landscape from cutaneous melanoma.1 BRCA1 and BRCA2 mutations appear in about 3%–16% of AMs.2,3 Pharmacologic inhibition of the DNA repair enzyme PARP has been approved by the FDA as monotherapy in patients with deleterious germline BRCA1/2 mutated advanced ovarian cancer, and the usage has been expanded to metastatic breast cancer, pancreatic cancer, and prostate cancer with homologous recombination repair (HRR) gene defects.4 However, whether AM with BRCA mutations is also sensitive to PARP inhibition is unknown. We identified a stage Ⅳ AM patient with a germline BRCA1 frameshift mutation (BRAC1 G1384Nfs∗7) who was resistant to anti-PD1 therapy. Both patient-derived xenograft and cells (PDX/PDC) models from the same AM patient were established. PARP inhibitor olaparib significantly decreased cell proliferation and slowed tumor growth by increasing DNA double-strand breakage in AM cancer cells. Administration of olaparib to the patient achieved stable disease for 3 months. This study provides preclinical and clinical evidence that PARP inhibitors can slow tumor growth in BRCA1-mutant advanced acral melanoma.
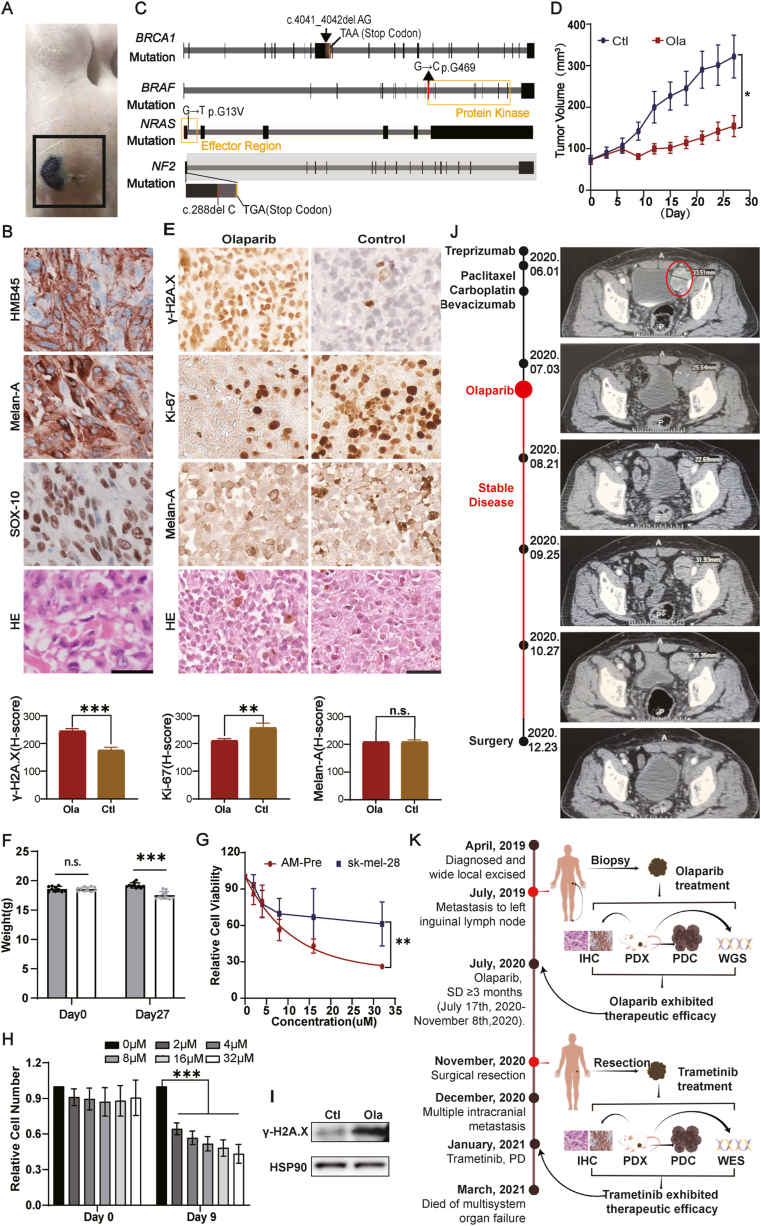
A 72-year-old man had a non-healing pigmented traumatic wound on his left planter (Fig. 1A) that was diagnosed as acral melanoma on March 21st, 2019. The patient underwent wide local excision (WLE) of the tumor at a nearby hospital on April 3rd, 2019, without TNM staging or adjuvant treatment. On July 24th, 2019, a lump at a left inguinal lymph node was identified and subsequently biopsied (Fig. S1). Immunohistochemically, the tissue presented high levels of HMB45, Melan-A, and SOX-10 (Fig. 1B). As a result, the patient was diagnosed with metastatic acral melanoma.
Figure 1.
Olaparib exhibited therapeutic efficacy in patient-derived xenograft and cell line (PDX and PDC) from an acral melanoma (AM) with BRCA1 germline mutation. (A) Image of AM primary lesion in the patient. (B) IHC staining of melanoma markers on the lymph node biopsy tissue. Scale bar = 50 μm. (C) Map of BRCA1, BRAF, NRAS, and NF1 mutations. (D) Efficacy of olaparib in PDX model derived from tumor biopsy prior to olaparib treatment (AM-Pre). Mice received 50 mg/kg olaparib twice a day. The mice treated with olaparib presented smaller tumor volumes. (E) IHC staining of PDX tumor tissues. After treatment, γ-H2A.X staining was increased while Ki67 staining was decreased. Scale bar = 50 μm. H-scores are shown below. (F) The mice treated with olaparib exhibited only marginal weight loss. (G) Comparison of olaparib sensitivity in AM-Pre cells and cutaneous melanoma cells sk-mel-28 by cell viability measurement. (H) Cell number changes after olaparib treatment in the AM-Pre PDC cells. Olaparib significantly inhibited the proliferation of AM-Pre cells. (I) Increased level of γ-H2A.X after olaparib treatment in PDX tumor tissue shown by Western blot. (J) The tumor size at the lymph node during each treatment process was evaluated by a CT scan. (K) Summary of the entire treatment decision process for the AM patient (By Figdraw (www.figdraw.com)). n.s., non-significant. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
The combination of anti-PD1 (toripalimab, 240 mg q14d) and antiangiogenic (apatinib, 250 mg qd) therapies was administered as first-line treatment, but apatinib was soon withdrawn because of uncontrollable severe hypertension (200/100 mmHg) while toripalimab was continued. Progressive disease (PD) was found on May 14th, 2020 with multiple enlarged and fused lymph nodes in the left groin and beside the left iliac vessels, with the largest diameter at 5 cm. The combination of albumin-bound paclitaxel (0.2 g d1 and d8), carboplatin (500 mg d1), and bevacizumab (400 mg d1) was administrated as second-line treatment. However, the patient could not tolerate the side effects including severe oral mucositis, thrombocytopenia, fever, fatigue, and joint soreness, and urged for alternative administration.
To explore other options for this case, targeted next-generation sequencing was conducted utilizing tumor tissue from lymph node biopsy. A previously unreported germline BRCA1 frameshift mutation causing a premature stop codon (G1384Nfs∗7) was found in this patient. Weak MAPK pathway activation mutations, including BRAF G469A, NF2 frameshift mutation (L97Wfs∗26), and NRAS G13V mutations were also detected (Table S1 and Fig. 1C).
Olaparib, an oral PARP inhibitor, has been approved by the FDA as monotherapy in patients with deleterious germline BRCA1 mutated advanced ovarian cancer, breast cancer, pancreatic cancer, and prostate cancer. Its risk of severe adverse events is low.4 Therefore, we reasoned that this AM with a germline BRCA1 frameshift mutation might also respond to olaparib with durable adverse effects.
Since there were no publications regarding olaparib treatment in AM patients, we first evaluated the efficacy as well as the toxicity of olaparib by carrying out a series of pre-clinical experiments in both PDX and PDC (AM-Pre) models from the same AM patient. Olaparib was administrated orally at 50 mg/kg twice a day in the PDX model. Significant tumor growth inhibition was observed after olaparib treatment (Fig. 1D). The tumors from the PDX models were subjected to immunohistochemical staining to further evaluate the drug efficacy. After being treated with olaparib, increased γ-H2A.X accumulation and decreased Ki-67 expression were observed, indicating increased DNA double-strand breakage and decreased proliferation in AM cancer cells (Fig. 1E). Mice receiving this dose of olaparib exhibited only marginal weight loss, consistent with previous observations (Fig. 1F). Meanwhile, the PDC, which was derived from the PDX tumor, also showed decreased cell viability as well as increased γ-H2A.X accumulation after olaparib treatment (Fig. 1G–I). Therefore, the administration of olaparib presented significant efficacy and mild adverse effects in the PDX model established from the patient's tumor tissue, encouraging the usage of olaparib in this AM patient.
The patient was fully educated and risk-informed, signed the informed consent, and then received olaparib monotherapy (300 mg qd + 150 mg qn). The treatment continued for a month, and CT scanning showed shrinking lesions in the left iliac fossa and inguinal area, and the patient was assessed as SD according to RECIST 1.1. The treatment was maintained for another month. Although CT scanning then showed PD (progressive disease), increasing the dosage of olaparib (300 mg bid) controlled the disease into SD again (Fig. 1J; Fig. S2A). Oral olaparib therapy was continued. Mild fatigue was the main adverse effect (Fig. S2B).
Because SD was achieved and no other metastatic lesions were observed in whole-body CT scanning, surgical resection was performed after full consideration of the patient's preference (Fig. 1J; Fig. S2C). In total, the administration of olaparib controlled the disease for more than 3 months, from July 17th, 2020, to November 8th, 2020.
For this AM patient, old age, ulceration, and distant lymph node metastasis implied high risks of distant metastasis, tumor recurrence, and poor prognosis, and post-olaparib treated (AM-Post) cells were less responsive to olaparib compared to AM-Pre cells (Fig. S3A). We proactively investigated other optimal treatment options in case this patient relapsed.
The AM tumor profiled here presented three weak MAPK activating mutations – BRAF G469A, single allelic NF2 loss, and NRAS G13V. These “weak activators” can in aggregate result in the MAPK pathway activating effects.5 Thus, MEK inhibitor trametinib was then evaluated as a rational treatment option for this AM patient.4 To determine the efficacy of trametinib on melanoma that had been exposed to olaparib, we first established AM-Post PDX and PDC models. By whole-exome sequencing (WES) of the PDX tissue, the same mutations of BRAF, NF2, and NRAS (Table S2) were detected in AM-Post as in AM-Pre. Trametinib was administered at 30 mg/kg orally once daily in mice for 15 days. Significant tumor regression was observed after trametinib treatment (Fig. S3B) and coupled to marginal weight loss (Fig. S3C). IHC staining of the tumor section showed reduced expression of p-ERK, Ki-67, and γ-H2A.X (Fig. S3D). 1 μM trametinib was sufficient to inhibit the proliferation and decrease the viability of AM cells (Fig. S3E, F). As a result, trametinib was viewed as the prioritized treatment option for tumor recurrence in this AM patient.
The patient was diagnosed with AM multiple intracranial recurrence 1.5 months after surgery (Fig. S3G). Owing to the COVID-19 pandemic, the patient was unable to be hospitalized for systemic treatment and refused whole-brain radiotherapy. On January 10th, 2021, the patient started trametinib monotherapy (2 mg qd) for 15 days. Unfortunately, the patient had a worsening headache and became unconscious soon afterward. Finally, the patient died of multisystem organ failure on March 4th, 2021. The entire treatment process is summarized in Figure 1K.
In conclusion, the PARP inhibitor olaparib induced SD for more than 3 months in one AM patient with BRCA1 germline mutation. The MEK inhibitor trametinib showed anti-tumor effects in PDC and PDX models of AM with combined weakly activating mutations of the MAPK pathway, but no clinical benefit was observed in this AM patient after the reoccurrence of intracranial metastasis. Our data should encourage further exploration of both strategies for treating AM patients with genetic deficiencies in DNA damage repair or multiple weak MAPK activation mutations, respectively.
Ethics approval and consent to participate
All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of the Third People's Hospital of Zhengzhou (No. 2020-04-004-M01 and 2020-04-006-M01). Informed consent was obtained from all subjects involved in the study. Written informed consent was obtained from the patient to publish this case.
Author contributions
Ruixin Jiang: formal analysis, investigation, project administration, writing - original draft. Xianbin Liang: formal analysis, resources, investigation. Ye Tao: formal analysis, methodology. Ronghui Xia: methodology. Ming Lei: funding acquisition, writing - review & editing. Bin Jiang: data curation, resources. Robert L Judson-Torres: conceptualization, writing - review & editing. Yanjie Zhang: data curation, supervision, writing - review & editing. Weizhen Zhang: methodology, resources, supervision, writing - review & editing. Hanlin Zeng: conceptualization, data curation, supervision, writing - review & editing.
Funding
This research was funded by the National Natural Science Foundation of China (No. 82204421, Hanlin Zeng); the Clinical Research Program of 9th People's Hospital, Shanghai Jiao Tong University School of Medicine (No. JYLJ201917, Yanjie Zhang); the Innovative Research Team of High-Level Local Universities in Shanghai, China (No. SHSMU-ZLCX20211700, Ming Lei and Hanlin Zeng).
Conflict of interests
The authors declare no conflict of interests.
Acknowledgements
We thank the support from colleagues of Zeng Lab and Zhang Lab of the Shanghai Institute of Precision of Medicine for scientific discussion. We thank Shufang He and Hong Lu of the Bioimaging Facility in Shanghai Institute of Precision Medicine for providing technical support and assistance in data collection.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.11.014.
Contributor Information
Yanjie Zhang, Email: Zhangyanjie@shsmu.edu.cn.
Weizhen Zhang, Email: lixiangx@henu.edu.cn.
Hanlin Zeng, Email: hanlin.zeng@shsmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Multimedia component 1
Multimedia component 2
Multimedia component 3
Multimedia component 4
References
- 1.Hayward N.K., Wilmott J.S., Waddell N., et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545(7653):175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 2.Yeh I., Jorgenson E., Shen L., et al. Targeted genomic profiling of acral melanoma. J Natl Cancer Inst. 2019;111(10):1068–1077. doi: 10.1093/jnci/djz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byeon S., Cho H.J., Jang K.T., et al. Molecular profiling of Asian patients with advanced melanoma receiving check-point inhibitor treatment. ESMO Open. 2021;6(1) doi: 10.1016/j.esmoop.2020.100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.U.S. Food & Drug Administration. Drugs@FDA: FDA-approved drugs. Accessed December 1, 2021. https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm.
- 5.Shain A.H., Bastian B.C. From melanocytes to melanomas. Nat Rev Cancer. 2016;16(6):345–358. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1
Multimedia component 2
Multimedia component 3
Multimedia component 4