Figure 3.
TF circuits that govern dedifferentiated liposarcoma programs
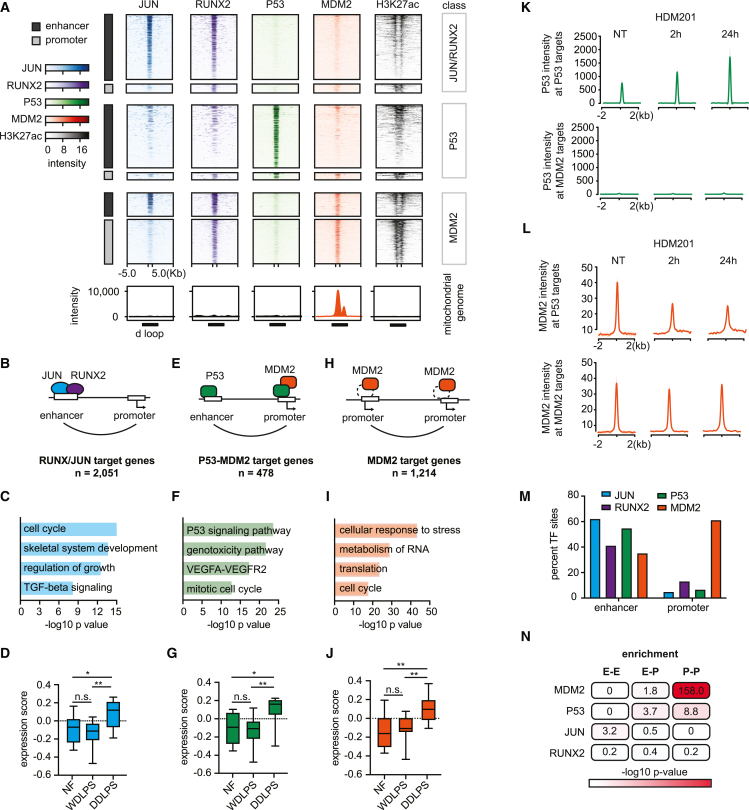
(A) Heatmap depicts TF binding (color heat) and H3K27ac (grayscale) over candidate enhancers and promoters (rows) of the LPS141 liposarcoma line. Peaks are binned into groups based on combinatorial TF binding, including JUN/RUNX peaks (n = 36,151), P53 peaks (n = 857), and MDM2 peaks without P53 (n = 1,544). Bottom boxes show TF binding on a linearized track of the circular mitochondrial genome centered on the regulatory d loop (black box).
(B, E, and H) Schematics summarize characteristic binding and looping patterns for JUN (blue), RUNX2 (purple), P53 (green), and MDM2 (orange).
(C, F, and I) Bars highlight gene sets enriched among JUN/RUNX2 (blue), P53 (green), and MDM2 (orange) targets, as defined by combination of ChIP-seq and HiChIP.
(D, G, and J) Box plots show expression of TF target genes (defined as in B, E, and H) in normal fat, WDLPS, and DDLPS samples. Two-tailed t test p values: (D) NF vs. DDLPS = 0.017, WDLPS vs. DDLPS = 0.0016; (G) NF vs. DDLPS = 0.012, WDLPS vs. DDLPS = 0.002; (J) NF vs. DDLPS = 0.007, WDLPS vs. DDLPS = 0.002.
(K) Aggregate plots show a P53 binding signal at P53 (top) or MDM2 (bottom) target sites in LPS853 cells with no treatment (NT) or after 2 h or 24 h of HDM201 treatment.
(L) Aggregate plots show MDM2 binding signal at P53 (top) or MDM2 (bottom) target sites in LPS853 cells with no treatment (NT) or after 2 h or 24 h of HDM201 treatment.
(M) Bar plot shows the proportion of TF binding sites that coincide with putative enhancers or promoters.
(N) Table depicts the degree of overlap between TF binding, per ChIP-seq, and sites engaged in enhancer-enhancer (E-E), enhancer-promoter (E-P), or promoter-promoter (P-P) loops, per HiChIP. Red heat represents the significance of overlap between indicated TF and loop pattern calculated with Fisher’s exact test using genome-wide loop patterns as background.