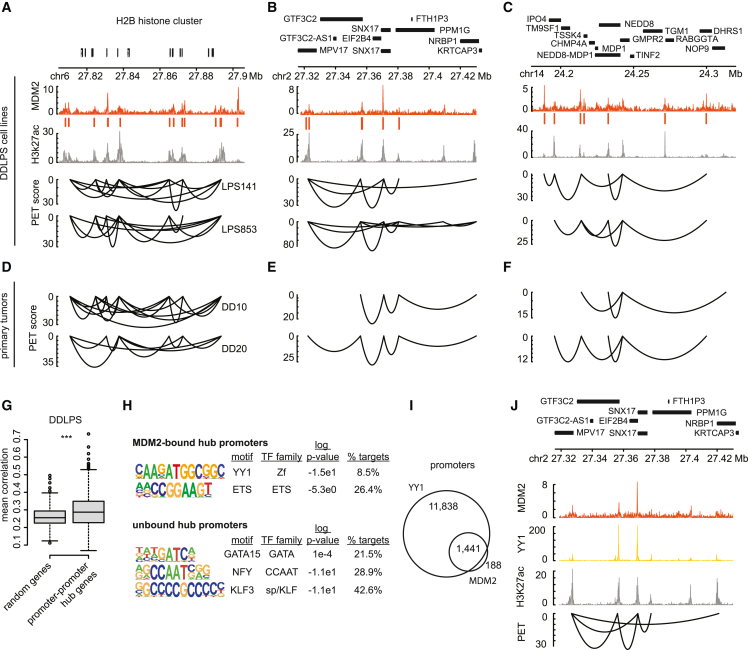
Figure 4.
Supraphysiologic MDM2 associates with promoter hubs
(A–C) Genomic tracks for three genomic regions with representative multi-way promoter hubs. Top tracks represent MDM2 binding intensity and called peaks (orange), and H3K27ac intensity (gray) in LPS141 cells. Bottom tracks depict HiChIP promoter-promoter loops in LPS141 and LPS853 cells. Loop height is proportional to the paired-end tag (PET) score. Genes are shown above.
(D–F) HiChIP promoter-promoter loops in two dedifferentiated tumors (DD10 and DD20) shown for the same loci.
(G) Box plots depict the correlation of the expression of promoter hub genes, compared with random gene sets in DDLPS tumors (n = 60) in the TCGA; two-tailed t test p value = 4.173e−5.
(H) Transcription factor binding motifs that are over-represented in MDM2-bound hub promoters (top) or unbound hub promoters (bottom).
(I) Venn diagram depicting overlap between YY1- and MDM2-bound promoters.
(J) Genomic track for representative promoter hub in the LPS853 cell line. Tracks represent, from top to bottom, MDM2 binding intensity (orange), YY1 binding intensity (yellow), and H3K27ac intensity (gray), and paired-end tag (PET) scores for promoter-promoter loops in H3K27ac HiChIP.