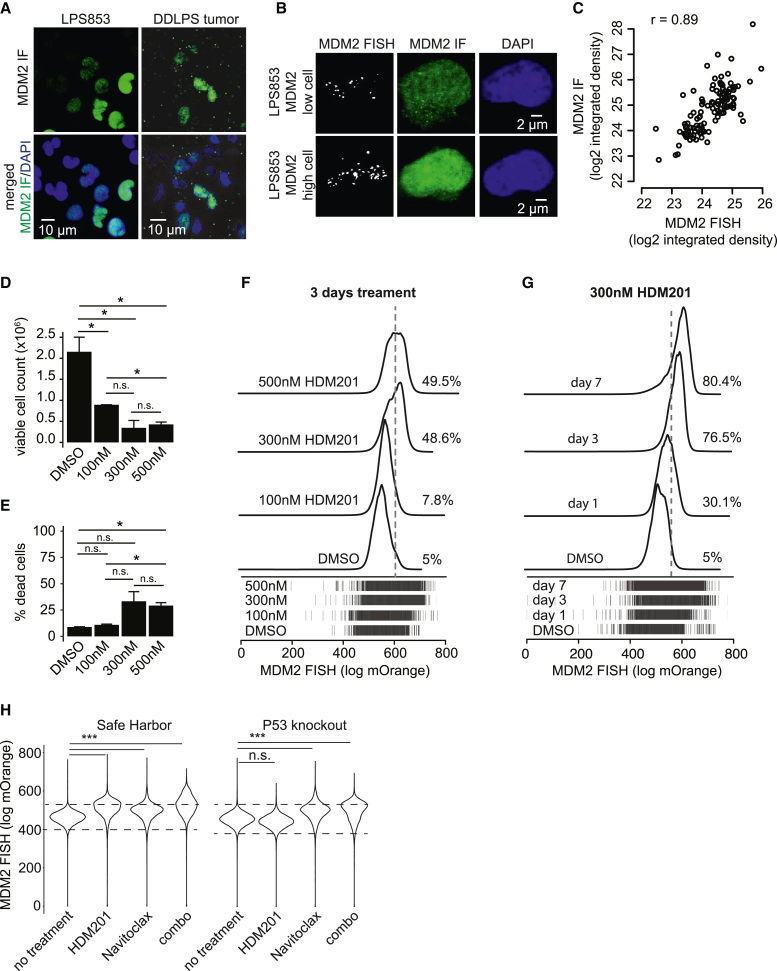
Figure 6.
Heterogeneous distribution of MDM2 copy number and expression confound efficacy of targeted therapies
(A) Representative 25× confocal IF images show MDM2 (green) and DAPI (blue) in LPS853 cells and a DDLPS tumor.
(B) Representative maximum-projection 63× confocal z stacks show FISH for the MDM2 locus (gray), IF for MDM2 protein (green), and DAPI (blue).
(C) Scatterplot compares MDM2 FISH and MDM2 IF intensity across 115 single cells.
(D and E) Bar plots comparing counts of viable (D) and dead (E) LPS853 cells following 3 days of HDM201 treatment.
(F) Histograms of MDM2 FISH intensity in LPS853 cells following 3 days of HDM201 treatment (n = 30,000 cells per condition). Dashed line represents the top 5% of the DMSO control. All drug-treated comparisons with the DMSO control displayed two-tailed t test p values < 2.2e−16.
(G) Histograms of MDM2 FISH intensity in LPS853 cells following a time course of 300 nM HDM201 treatment (n = 35,000 cells per condition). Dashed line represents the top 5% of the DMSO control. All drug-treated comparisons with the DMSO control displayed two-tailed t test p values < 2.2e−16.
(H) Violin plots show distribution of MDM2 FISH for safe harbor and P53 knockout LPS853 cells following 3 days treatment with DMSO (control), 100 nM Navitoclax or 300 nM HDM201 as single agents or in combination (n = 30,000 cells per condition). Dashed lines represent the top 5% and bottom 5% of the DMSO control. All comparisons marked as significant displayed two-tailed t test p values < 2.2e−16.