Figure 5.
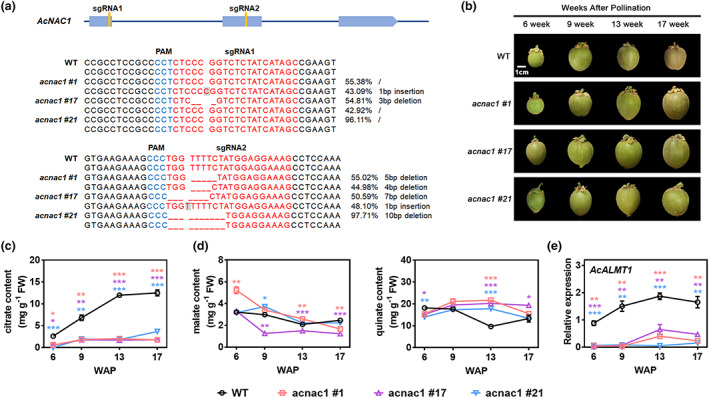
CRISPR‐Cas9 engineered AcNAC1 mutations result in reduced fruit citrate content. (a) Position of two target sites in AcNAC1 and Hi‐TOM sequencing of the CRISPR edited sites in RF‘DH’ acnac1 line #1, #17 and #21. sgRNA‐targeted sequences are highlighted in red and a protospacer adjacent motif (PAM) are shown in blue. Deletion and insertion are represented by dashed line and in grey background, respectively. Each mutation type is represented by the relative percentages. Mutation reads lower than 10% were filtered during data analysis. (b) Four developmental stages of WT (RF‘DH’) and three independent RF‘DH’ acnac1 lines #1, #17 and #21. WAP, weeks after pollination. Bar = 1 cm. (c) Dynamic profiles of citrate during four developmental stages of WT and the AcNAC1 CRIPSR/Cas9 kiwifruits. (d) Dynamic profiles of malate and quinate during four developmental stages of WT and the AcNAC1 CRIPSR/Cas9 kiwifruits. (e) The expression patterns of AcALMT1 in WT and the AcNAC1 CRIPSR/Cas9 kiwifruits. Data are presented as means ± SE of three independent biological replicates. Asterisks above bars indicate significant difference from the WT (*P < 0.05; **P < 0.01; ***P < 0.001) analysed by the Student's t‐test.
