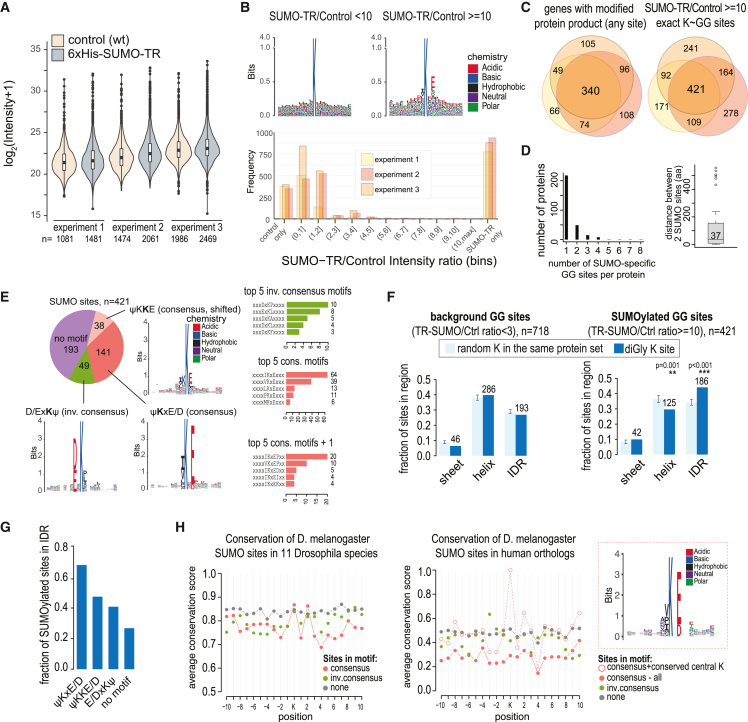
Figure 2.
Characteristics of the ovarian SUMOylome
(A) Distribution of diGly site intensities in different samples.
(B) (Top) Amino acid frequencies flanking predicted K-ε-GG, for sites with high and low SUMO-TR:control intensity ratios in a representative replicate. Also see Figure S1. (Bottom) Distribution of SUMO-TR:Control intensity ratios of diGly sites in each experiment.
(C) Overlap of exact SUMO sites or SUMO-modified genes (irrespective of exact sites) between replicates.
(D) Numbers of predicted SUMO sites per protein and distance between sites for proteins with two sites.
(E) Sequence motifs at 421 high-confidence SUMO sites. The predicted diGly-modified lysine is in bold. Bar graphs show the counts of topmost frequent amino acids at positions −1 and +2, and −2, −1, and +2 from the modified lysine.
(F) Localization of diGly sites with high or low intensity ratios in predicted structured regions or IDRs, compared with randomly selected lysines from the same protein over 1,000 iterations. Error bars represent SD.
(G) Localization of diGly SUMO sites embedded in different motifs to IDRs.
(H) Conservation of predicted SUMO sites and flanking regions embedded in indicated motifs in 11 other Drosophila species and human. Sequence logo shows the amino acid frequencies of homologous regions in human proteins, where the Drosophila site is embedded in a consensus SUMOylation motif and the central lysine is conserved between fly and human.