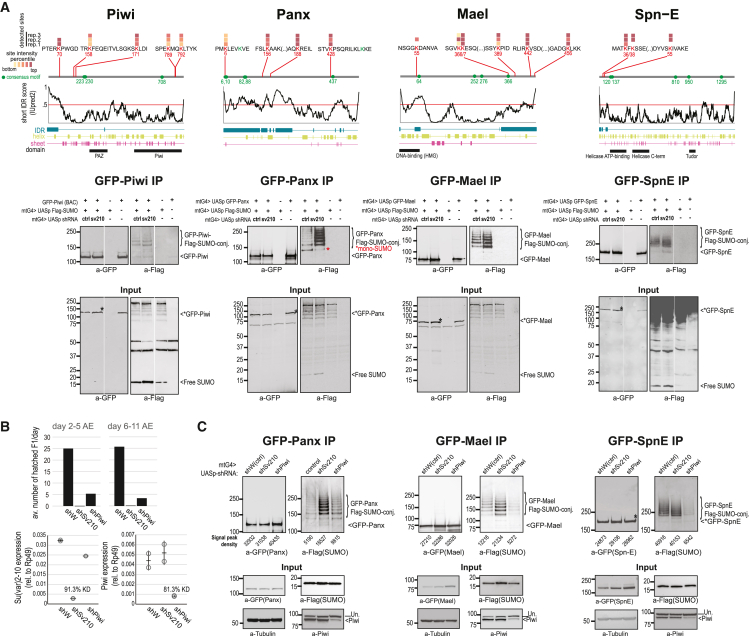
Figure 5.
SUMOylation of Piwi, Mael, Spn-E, and Panx
(A) (Top) Diagrams of structural features, consensus SUMOylation motifs, and experimentally detected diGly sites with TR-SUMO:control intensity ratios >10. Color indicates diGly site intensity percentiles. (Bottom) WB analysis of target SUMOylation in the female germline. Ovary lysates from flies carrying indicated transgenes (mtG4 = maternal tubulin Gal4 driver) were subjected to GFP immunoprecipitation (IP) followed by WB detection first with anti-FLAG antibody and, after stripping, anti-GFP antibody. Note that SUMOylated species are only detectable by the highly sensitive anti-FLAG antibody, but not anti-GFP antibody. Lanes in Piwi and Spn-E gels were cropped and flipped for sample order consistency. Images are representative of three or four independent experiments per target.
(B) Efficiency of su(var)2-10 and piwi germline knockdown estimated by fertility test (top) and RT-qPCR (bottom). Results are from samples from the same genetic crosses used for WB in (C), GFP-Panx. Error bars represent SD.
(C) WB of target SUMOylation in the female germline upon white (control), su(var)2-10, and piwi germline knockdown. Images are representative of two independent experiments. Un., unspecific band.