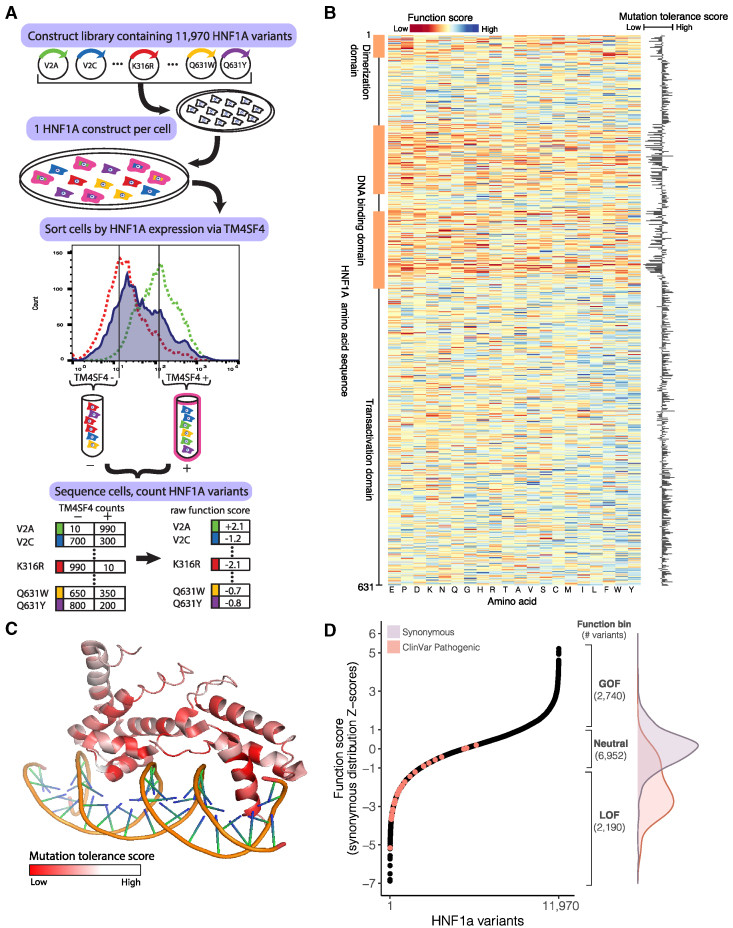
Figure 1.
Identification of GOF variants from comprehensive functional testing of 11,970 HNF1A variants in human hepatocytes
(A) A library of 11,970 HNF1A constructs was synthesized, with each construct encoding a single amino acid substitution. The construct library was introduced into HUH7 hepatocytes (deleted for endogenous HNF1A) at a dilution of one construct per cell. The resulting polyclonal population of HUH7 hepatocytes was separated via FACS according to the expression of the known HNF1A transcriptional target TM4SF4 and sorted into low (−) and high (+) bins of HNF1A activity. Activity cutoffs were established through flow cytometry experiments of HNF1A KO cells (dashed red line) and WT cells (dashed green line). Each bin of cells was sequenced at the transgenic HNF1A locus to identify and tabulate the introduced variants. Each HNF1A variant was assigned a function score based on its abundance in the low and high TM4SF4 expression bins.
(B) Heatmap of 11,970 HNF1A variant function scores, arranged according to the primary amino acid sequence (rows). Function scores lower than WT are shaded red, and function scores greater than WT are shaded blue. Function scores averaged (mean) at each amino acid position are plotted to the right, showing the level of tolerance for any amino acid substitution away from WT at each position.
(C) Mutation tolerance scores as described in (B) overlaid on the crystalized protein structure of HNF1A DNA-binding domain (PF04814). Positions intolerant of amino acid changes (i.e., lower function scores) are shaded red. Helices that make direct contacts with the DNA are the most intolerant of mutations.
(D) (Left panel) HNF1A function scores ranked for all 11,970 amino acid variants tested, and ClinVar-annotated pathogenic variants (n = 29) are highlighted in red. (Right panel) Function bins correspond to variants with function scores above (GOF), within (neutral), or below (LOF) ± 1 Z-score of the synonymous distribution, shown with the total number of variants per bin. Overlaid are the function score distributions of the 613 synonymous HNF1A variants tested (purple) and the 29 ClinVar pathogenic variants (red).