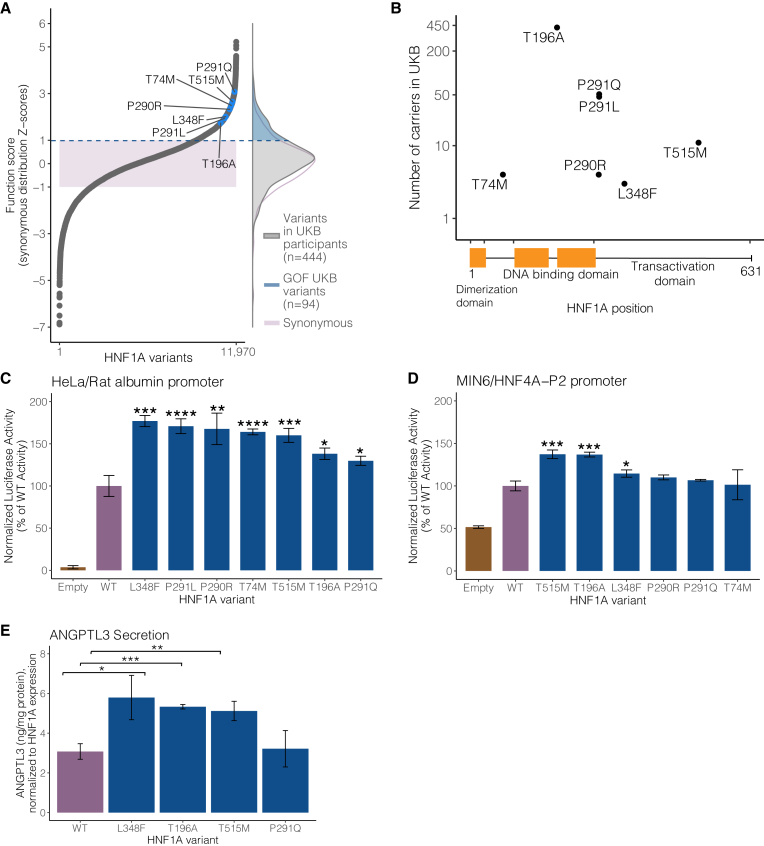
Figure 2.
GOF HNF1A variants are carried in the general population and increase transcriptional activity in multiple cellular contexts
(A) (Left panel) Selected human HNF1A variants with function scores greater than WT highlighted in blue among the full distribution of all 11,970 amino acid variants tested. The shaded purple box represents ± 1 Z-score of the distribution of 613 synonymous HNF1A variants tested. (Right panel) The distributions of function scores of rare protein-coding HNF1A variants (n = 444) identified from 454,756 sequenced individuals in the UK Biobank (UKB) and of the 613 synonymous variants. An upper tail of function-increasing variants in UKB individuals is highlighted in blue (n = 94 unique variants in 1,469 individuals).
(B) Location of top-scoring variants selected for validation along the HNF1A protein primary amino acid sequence with number of human variant carriers identified in 454,756 exome sequences from the UKB.
(C) Putative GOF variants were individually recreated and tested for transcriptional activation using luciferase reporters in HeLa cells (which lack endogenous HNF1A activity) on the rat albumin promoter. Activity measurements are shown as percentages of WT HNF1A activity ± SEM; n = 3. Basal promoter activity in cells is measured by transfection of vectors lacking the HNF1A transgene (empty) (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, linear mixed model).
(D) The same variants described in (B) were tested for transcriptional activity in mouse insulinoma MIN6 cells on the HNF4A-P2 promoter.
(E) GOF HNF1A variant constructs were transfected into HNF1A-deleted HUH7 hepatocytes, and ANGPTL3 levels were measured by ELISA 48 h post-confluency (n = 3 per variant). Transfection with HNF1A p.L348F, p.T196A, and p.T515M increases ANGPTL3 secretion compared with WT (∗∗∗p < 0.0005,∗∗p < 0.005, ∗p < 0.05, linear mixed model).