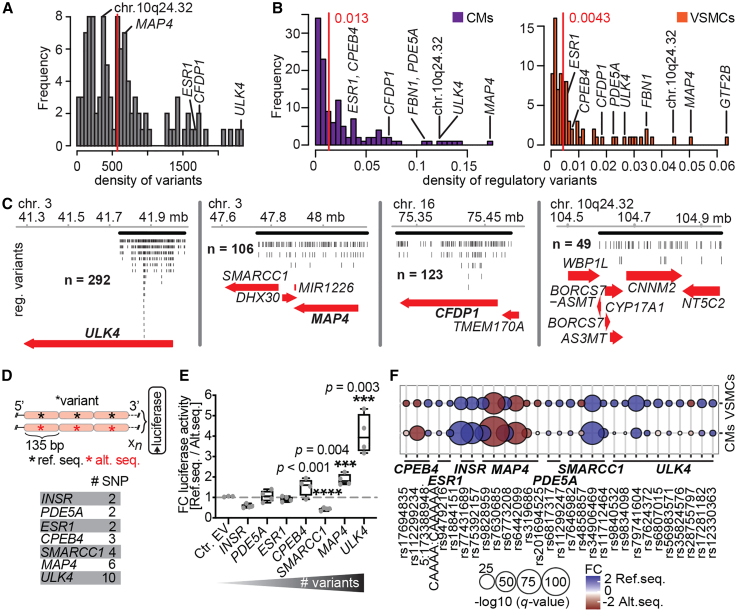
Figure 4.
GWAS loci can have high densities of regulatory variants
(A) Density plot of the number of variants in LD per GWAS variant per megabase. Red line denotes median (572 variants/Mb), genes with high number of regulatory variants (B) are labeled.
(B) Density plot of the ratio between the number of regulatory variants in LD per GWAS variant and the number of variants per megabase in each LD block in CMs (left) and VSMCs (right). Red lines denote medians.
(C) Genome browser examples with numbers of aggregated regulatory variants at ULK4, MAP4, CFDP1, and 10q24.32 loci. Black lines indicate LD blocks.
(D) Scheme for luciferase assays. Reference and alternative sequences of every haplotype of concatenated regulatory alleles were analyzed in the 5′ → 3′ direction 5′ of luciferase. Number of variants concatenated for luciferase assays per locus is shown.
(E) Fold changes in luciferase activity comparing haplotypes of reference vs. alternative sequences to empty vector (four biological replicates). Asterisks show significance determined by unpaired two-tailed t tests (∗∗∗p < 0.004, ∗∗∗∗p < 0.0001). These comparisons are considered to be independent. Therefore, multiple-testing correction was not applied.
(F) Bubble plot showing the MPRA regulatory capacity of variants tested in the luciferase assay. See also Figure S4; Tables S7, S8, S9, S10, S11, S12, S13, and S14.