Figure 5.
Regulatory variants spatially converge with trait-relevant genes in 3D genome organization
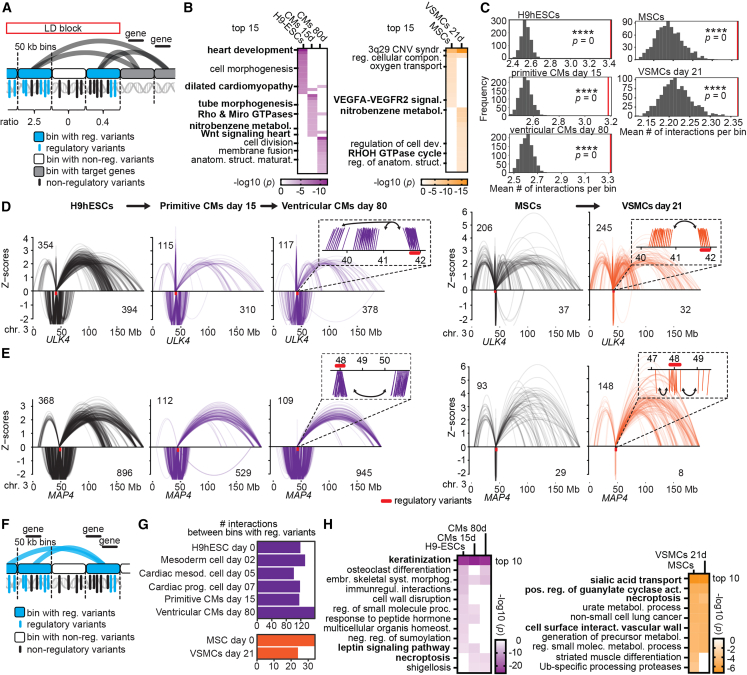
(A) Scheme of Hi-C/Omni-C analysis with genome binned in 50 kb bins to analyze significant interactions of bins harboring regulatory variants with target genes. Ratios of regulatory over non-regulatory variants of each bin were generated to account for variant distribution per bin.
(B) Enrichment analysis of genes within 50 kb bins of significantly interacting bins with at least one regulatory variant. Cutoff p <0.001 to reduce number of genes for enrichment analysis. Terms listed are related to cardiovascular function according to the literature; BP-related terms are highlighted in bold.
(C) Number of genomic interactions for bins with regulatory variants compared with 1,000× permuted random sets of interactions. Red line denotes mean number of interactions per bin, asterisks denote significance (∗∗∗∗empirical p = 0).
(D and E) Arch plots (10% opacity) across entire chromosomes showing all significant genomic contacts of bins (50 kb) with regulatory variants at (D) ULK4 and (E) MAP4 in three Hi-C and in two Omni-C datasets. Numbers in arch plots indicate total number of interactions. Z scores indicate interactions occurring more (positive) or less (negative) often than expected considering the genomic linear distance. Zoomed-in windows exemplify that consecutive bins interact with the same targets.
(F) Scheme of Hi-C/Omni-C analysis to determine solely genomic contacts between bins harboring regulatory variants (reciprocal interactions).
(G) Number of reciprocal interactions per cell type.
(H) Enrichment analysis of genes involved in reciprocal interactions revealed specific cardiovascular pathways (highlighted in bold font). See also Figures S5 and S6; Table S15.