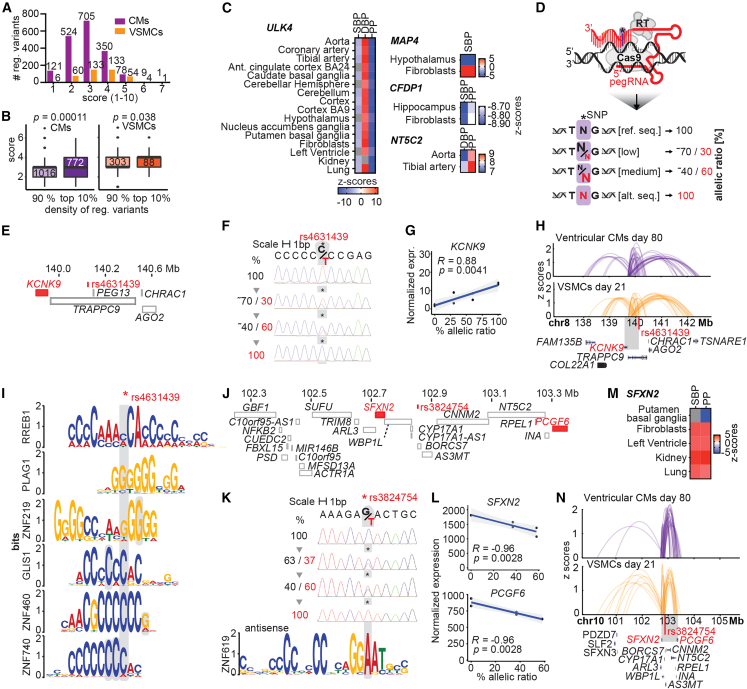
Figure 6.
Heuristic scoring identifies likely causal variants, and CRISPR prime editing definitively determines variant target genes
(A) Distribution of heuristic scores for all regulatory variants. Numbers above bars indicate numbers of regulatory variants per score.
(B) Comparison of the heuristic ranking scores between loci with high regulatory variants density (top 10%) and low variant density (bottom 10%). Significance from Mann Whitney test, CMs: p = 0.00011, VSMCs: p = 0.038. Number of variants under each category is shown inside each box.
(C) Significant associations between genetically predicted gene expression (GPGE) of ULK4, MAP4, CFDP1, and NT5C2 genes with SBP, DBP, and PP (pulse pressure) traits in BP-relevant tissues.
(D) CRISPR prime editing (PE) comprising Cas9 nickase, reverse transcriptase (RT), and pegRNAs precisely edited regulatory variants at allelic ratios: 100% reference sequence, low editing (∼30% of reference sequence), medium editing (∼60% of reference sequence), and ∼100% alternative sequence.
(E) Gene model for variant rs4631439 determining KCNK9.
(F) PE efficiencies (%) shown in Sanger sequencing tracks of rs4631439.
(G) Correlation between KCNK9 gene expression and PE allelic ratios for rs4631439.
(H) Arch plots (25% opacity) show genomic interactions as Z scores of locus harboring rs4631439 (in 50 kb bin) in Hi-C data of ventricular CMs (day 80) and in Omni-C data of VSMCs (day 21).
(I) Selection of predicted TFBSs that are disrupted at the variant’s position. Ambiguous mapping of TF motifs to multiple positions around variants is highlighted in gray.
(J) Gene model for variant rs3824754, SFXN2, and PCGF6.
(K) PE efficiencies (%) of rs4631439 and predicted TFBSs for ZNF619 that get disrupted at the variant’s position.
(L) Correlations between gene expression for SFXN2 and PCGF6 and rs3824754 allelic ratios from PE.
(M) Significant associations between GPGE of SFXN2 with SBP and PP traits in BP-relevant tissues.
(N) Arch plots (25% opacity) show genomic interactions as Z scores of locus harboring rs3824754 (in 50 kb bin) with SFXN2 and PCGF6 in Hi-C/Omni-C data. See also Figures S5 and S6; Tables S17, S18, S19, S20, S21, and S22.