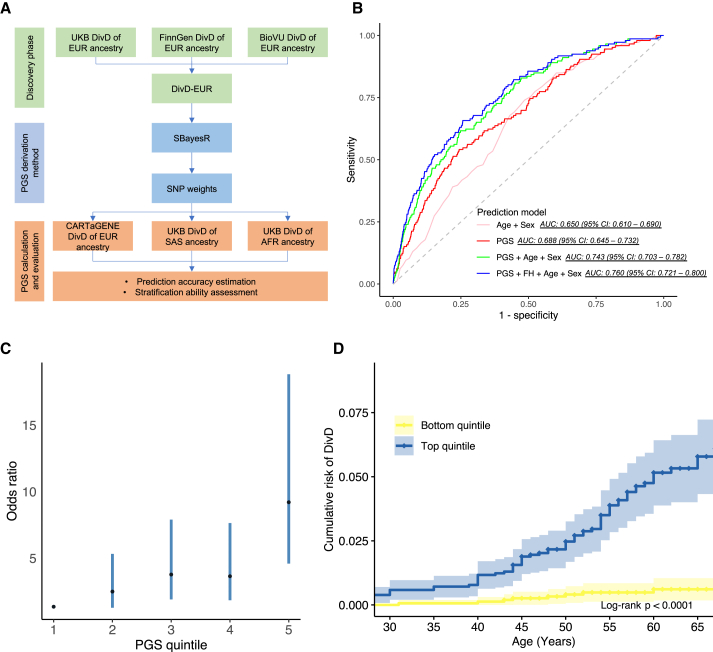
Figure 5.
DivD PGS prediction analyses
(A) Workflow for prediction analyses. SBayesR was applied to derive the SNP weights for downstream PGS calculation together with DivD-EUR GWAS summary statistics.
(B) AUC of DivD risk prediction models in CARTaGENE cohort. The derived PGS provides comparable predictive ability on top of traditional risk factors (age and sex). The AUC is based on a logistic regression model with coefficients estimated for age, sex, family history (FH), and PGS estimated from the CARTaGENE data.
(C) OR for developing DivD in CARTaGENE cohort (146 individuals with DivD and 7,550 controls) for each PGS quintile. The black dots are the OR values and the error bars are the 95% CIs.
(D) The cumulative risk of DivD for individuals in the top and bottom quintile (with 95% CIs) of PGS of the CARTaGENE data.