Figure 1.
Overview of the OpenPBTA project
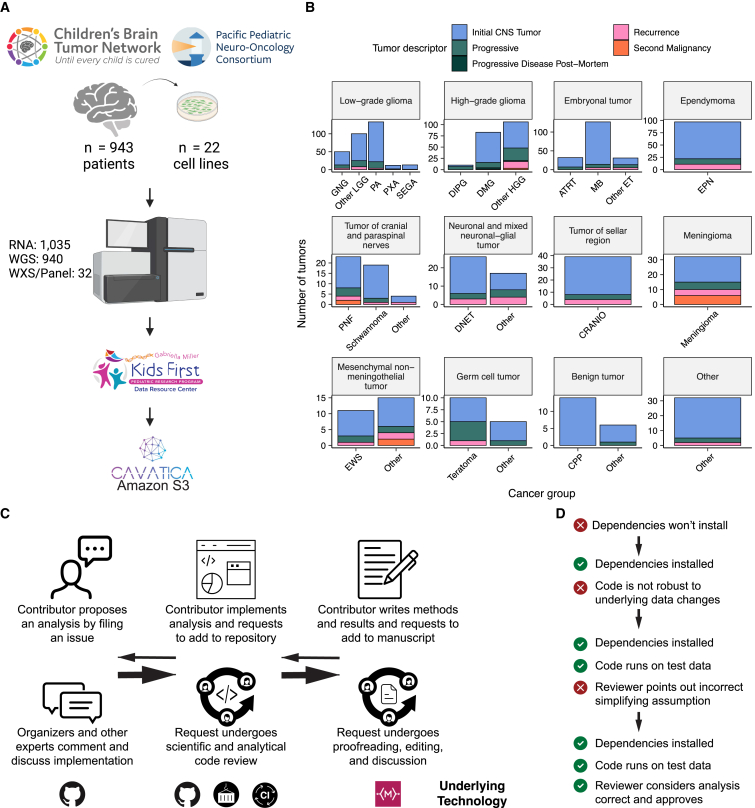
(A) CBTN and PNOC collected tumors from 943 patients. 22 tumor cell lines were created, and over 2,000 specimens were sequenced (n = 1035 RNA-seq, n = 940 WGS, and n = 32 WXS or targeted panel). The Kids First Data Resource Center Data harmonized the data using Amazon S3 through CAVATICA. Panel created with BioRender.com.
(B) Number of biospecimens across phases of therapy, with one broad histology per panel. Each bar denotes a cancer group (abbreviations: GNG, ganglioglioma; other LGG, other low-grade glioma; PA, pilocytic astrocytoma; PXA, pleomorphic xanthoastrocytoma; SEGA, subependymal giant cell astrocytoma; DIPG, diffuse intrinsic pontine glioma; DMG, diffuse midline glioma; other HGG, other high-grade glioma; ATRT, atypical teratoid rhabdoid tumor; MB, medulloblastoma; other ET, other embryonal tumor; EPN, ependymoma; PNF, plexiform neurofibroma; DNET, dysembryoplastic neuroepithelial tumor; CRANIO, craniopharyngioma; EWS, Ewing sarcoma; CPP, choroid plexus papilloma).
(C) Overview of the open analysis and manuscript contribution models. Contributors proposed analyses, implemented it in their fork, and filed a pull request (PR) with proposed changes. PRs underwent review for scientific rigor and accuracy. Container and continuous integration technologies ensured that all software dependencies were included and that code was not sensitive to underlying data changes. Finally, a contributor filed a PR documenting their methods and results to the Manubot-powered manuscript repository for review.
(D) A potential path for an analytical PR. Arrows indicate revisions.